
Towards Novel Methods for Effective Transfer Learning and
Unsupervised Deep Learning for Medical Image Analysis
Mijung Kim, Jasper Zuallaert and Wesley De Neve
Center for Biotech Data Science, Ghent University Global Campus, Songdo, Incheon, 305-701, Korea
IDLab, Ghent University - imec, Ghent, 9000, Belgium
{mijung.kim, jasper.zuallaert, wesley.deneve}@ugent.be
1 RESEARCH PROBLEM
1.1 Introduction
Thanks to computational and algorithmic advances,
as well as an increasing availability of vast amounts
of data, deep learning techniques have substantially
improved over the past decade (LeCun et al., 2015).
Specifically, in recent years, deep learning techniques
have been successfully applied to the field of im-
age analysis (Szegedy et al., 2016a), speech recog-
nition (Hinton et al., 2012), and natural language pro-
cessing (Mikolov et al., 2013), showing that they are
increasingly able to outperform traditional machine
learning approaches that typically rely on manual fea-
ture engineering. Furthermore, in collaboration with
healthcare institutes, companies like Google and IBM
have recently started with the application of deep
learning techniques to medical use cases. As an ex-
ample, the authors of (Wong and Bressler, 2016) dis-
cuss the usage of deep learning techniques for diag-
nosing diabetic retinopathy, an eye disease that occurs
when diabetes causes damage to the retina.
Compared to the application of conventional ma-
chine learning approaches to medical images, the ap-
plication of deep learning techniques remains chal-
lenging. Indeed, medical image sets are often lim-
ited in size and (partially) unlabeled (Chen and Lin,
2014), due to privacy concerns, acquisition difficul-
ties, and/or the time-consuming nature of manual la-
beling. However, when applying deep learning tech-
niques, the following rule of thumb usually holds
true: the more data that can be leveraged during train-
ing, the higher the effectiveness of prediction (LeCun
et al., 2015). As a result, given that it is difficult to get
access to vast collections of properly labeled medical
images, predictive models obtained through the us-
age of deep learning techniques typically suffer from
overfitting, thus leading to inaccurate diagnoses.
Restrictions in terms of size and labeling are not
limited to medical datasets; datasets in other applica-
tion areas are facing these challenges as well (Santoro
et al., 2016). Therefore, more and more research ef-
forts are dedicated to addressing these shortcomings.
One promising approach towards dealing with
small-sized datasets is the usage of transfer learn-
ing, a technique that can be used to improve a model
from one domain by leveraging knowledge from a
related domain. Compared to training from scratch
with small datasets, experimental analysis has demon-
strated that transfer learning may reduce the relative
error with up to 50% (Yosinski et al., 2014; Azizpour
et al., 2015). However, compared to training from
scratch with vast datasets, there is still significant
room for improvement (Szegedy et al., 2016a).
Another interesting approach towards dealing
with small-sized datasets, as well as with a lack of
labeled samples, is the usage of unsupervised deep
learning, which allows exposing structure and seman-
tics in unlabeled datasets. Indeed, several unsuper-
vised deep learning techniques have recently been in-
troduced, for instance making it possible to generate
similar images out of a given set of images.
Our doctoral research will focus on the construc-
tion and evaluation of new predictive models for med-
ical image diagnosis, through the development of
novel methods for effective transfer learning and un-
supervised deep learning, so to be able to overcome
limitations in terms of size and labeling. In the fol-
lowing section, we outline a number of relevant re-
search questions that we set out to answer.
1.2 Research Questions
Given the current state-of-the-art in the field of deep
learning, the main question of our doctoral research
has been set as follows:
”Given the availability of small-sized sets of
medical images, how can deep learning techniques
be leveraged for medical image analysis, obtaining a
high effectiveness of prediction without overfitting?”
32
Kim, M., Zuallaert, J. and Neve, W.
Towards Novel Methods for Effective Transfer Learning and Unsupervised Deep Learning for Medical Image Analysis.
In Doctoral Consortium (DCBIOSTEC 2017), pages 32-39

Using transfer learning as a starting point, we
can employ an additional technique that is com-
plementary in nature, called data augmentation.
When applying data augmentation to sets of medical
images, the idea is to generate additional training
images by for instance rotating, cropping, and/or
translating the original images (Krizhevsky et al.,
2012). Thus, to facilitate the effective application
of currently available deep learning techniques, or
modified versions thereof, to medical use cases, our
doctoral research will also try to answer the following
related questions:
• ”What (novel) transfer learning approaches work
well for medical image diagnosis? Why is it that
these transfer learning approaches work well?”
• ”What (novel) strategies towards fine-tuning of
pre-trained neural networks work well for med-
ical image understanding? Why is it that these
fine-tuning strategies work well?”
• ”Is data augmentation during training able to
help in improving the effectiveness of deep learn-
ing models that aim at medical image analysis?
Which (novel) methods for data augmentation can
be leveraged? Why is it that particular methods
for data augmentation work well?”
Finally, to deal with both small-sized and (par-
tially) unlabeled sets of medical images, we will also
explore approaches for unsupervised deep learning.
In this context, our doctoral research aims at finding
an answer to the question below:
”What (novel) unsupervised deep learning ap-
proaches are suitable for dealing with both small-
sized and unlabeled sets of medical images? Why is
it that these unsupervised deep learning approaches
work well?”
2 STATE-OF-THE-ART
In this section, we examine a couple of state-of-the-
art approaches related to transfer learning and unsu-
pervised deep learning. By having a close look at
these approaches, we are able to develop our own ap-
proaches towards overcoming challenges in the area
of deep learning-based medical image analysis.
2.1 Transfer Learning
Transfer learning is typically implemented by means
of the following two steps (Yosinski et al., 2014):
1. Given a task, train a source network on a source
dataset.
2. Given another task, transfer the learned features
to a target network for a particular target dataset.
The above two steps can be formally expressed as
follows (Pan and Yang, 2010):
”Given a source domain D
S
and a learning task
T
S
, a target domain D
T
and a learning task T
T
,
transfer learning aims at improving the learning
of the target prediction function f
T ()
in D
T
using
the knowledge in D
S
and T
S
, where D
S
6= D
T
, or
T
S
6= T
T
.”
In (Yosinski et al., 2014), the authors demonstrate
the high tranferability of a deep neural network, using
an AlexNet architecture trained on ImageNet. In do-
ing so, they make use of fine-tuning, a techniques that
adapts the pre-trained network to the target dataset
and task by adjusting the learned features, with the
goal of achieving a higher effectiveness. In particular,
the last layer of the network is replaced with a new
layer that takes into account the characteristics of the
target dataset (Girshick et al., 2014). The authors then
experiment with freezing different layers and retrain-
ing the remaining layers to find the best way to realize
transfer learning.
In summary, the authors of (Yosinski et al., 2014)
were able to make the following observations. First,
when the source and target datasets were similar,
transfer learning slightly outperformed a source net-
work by 0.02 in terms of top-1 accuracy. However,
when dissimilar datasets were fed to the network, the
effectiveness dropped by 0.10 in terms of top-1 ac-
curacy. The latter observation was also confirmed
by (Azizpour et al., 2015), illustrating that effective
transfer learning remains an open research challenge.
2.2 Unsupervised Deep Learning
A generative model is self-explanatory in nature, pro-
ducing samples that share similar features with sam-
ples available in a source dataset. Producing samples
is often done by making use of Markov Chain Monte
Carlo sampling, and Gibbs sampling in particular.
Proposed by Geoffrey Hinton in 1985 (Ackley et al.,
1985), Boltzmann Machines and derivative models
such as Restricted Boltzmann Machines, Deep Belief
Networks, and Deep Boltzmann Machines are repre-
sentative examples of deep generative models (Good-
fellow et al., 2016). As discussed in the next sections,
new approaches for sample generation have recently
been proposed, seeing their combination with deep
learning techniques.
Towards Novel Methods for Effective Transfer Learning and Unsupervised Deep Learning for Medical Image Analysis
33

Figure 1: Relation between the generator and the discrimi-
nator (Belghazi, 2016)).
2.2.1 Variational Autoencoders
The Variational Autoencoder (VAE) proposed
by (Kingma and Welling, 2013) has a structure that
is similar to the structure of the vanilla autoencoder
introduced in (Rumelhart et al., 1985). However,
the VAE is a stochastic model that makes use of
a probabilistic encoder q
φ
= (z|x) to approximate
the true posterior distribution p(z|x) of the latent
variables, where x is a discrete or continuous variable
and where z is an unobserved continuous random
variable. Due to the intractability of the posterior
distribution, the authors suggest the use of the
Stochastic Gradient Variational Bayes (SGVB) esti-
mator to approximate the true posterior distribution
of the latent variables. The SGVB estimator enables
backpropagation by adopting ε, where ε ∼ N (µ, σ)
and where z = µ + εσ, with µ denoting the mean and
σ the standard deviation.
By leveraging a stochastic graphical model with
a Bayesian network, VAEs have been successfully
used for the purpose of generating handwritten dig-
its (Kingma and Welling, 2013; Salimans et al., 2015)
and face images (Rezende et al., 2014).
2.2.2 Generative Adversarial Networks
A Generative Adversarial Network (GAN), as pro-
posed in (Goodfellow et al., 2014) and as visualized
in Figure 1, consists of two parts: a generator and a
discriminator. The generator produces new samples
similar to the real data that were fed into the network.
The newly produced samples are then judged by the
discriminator, to determine whether they are counter-
feit in nature or not. By repeating the training process,
the network is able to find an equilibrium for both.
A deep convolutional GAN (Radford et al., 2015),
typically abbreviated as DCGAN, also consists of a
generator and a discriminator. However, the sample
generation and discrimination processes are different.
In particular, in a DCGAN, the generator makes use
of deep convolutional networks, whereas the discrim-
inator is implemented by means of deconvolutions.
As discussed by (Frans, 2016), since VAEs fol-
low an encoding-decoding scheme, we can compare
the generated images directly to the original images,
something that is not possible to do with GANs.
Moreover, GANs are more difficult to optimize due
to unstable training dynamics (the generator and dis-
criminator sub-networks within a GAN are trained us-
ing opposed target functions). However, given that
VAEs use mean squared error instead of an adversar-
ial network, GAN images are currently more sharp
than VAE images. Indeed, GANs are able to detect
and thus reject blurry images.
Given the focus of GANs to learn to make images
that look real in general, the synthesized images tend
to combine features from different types of objects.
Two research efforts that aim at exercising more con-
trol over this behaviour are (Salimans et al., 2016)
and (Chen et al., 2016), and where both research ef-
forts add multiple objectives to the cost function of the
discriminator. Furthermore, research efforts have also
been dedicated to mitigating VAE bluriness, either by
making use of perceptual quality metrics (Dosovit-
skiy and Brox, 2016) or by making use of a recur-
rent generative autoencoder (Guttenberg et al., 2016).
Finally, it is interesting to point out that initial re-
search has also been done on combining VAEs and
GANs, using the same encoder-decoder configura-
tion, but leveraging an adversarial network as a metric
for training the decoder (Boesen et al., 2015).
3 OUTLINE OF OBJECTIVES
The main objective of our research is to construct
novel predictive models for medical image diagnosis.
In that regard, we plan to develop and evaluate novel
deep learning-based techniques that are complemen-
tary to already existing techniques, answering the re-
search questions formulated in Section 1.2. Particular
attention will be paid to the construction of novel pre-
dictive models that meet the following sub-objectives:
• Reliability - This is he most important factor
in medical use cases. Therefore, our research
will focus on obtaining high values for metrics
such as accuracy, sensitivity, and specificity, and
where these metrics are widely used in the field of
medical image analysis (Lalkhen and McCluskey,
2008). We discuss these metrics in more detail in
Section 4.2.
• Transferability - The newly developed predictive
models need to be transferable. This means that,
regardless of the dataset(s) they were trained on,
the predictive models will be applicable to other
data domains, while still producing reliable re-
sults. In other words, thanks to transferability,
DCBIOSTEC 2017 - Doctoral Consortium on Biomedical Engineering Systems and Technologies
34

our predictive models may not only be applied
within the same domain, but also across differ-
ent domains, and where these domains may also
come with small-sized data sets (e.g., from anal-
ysis of mammogram images to analysis of lung
X-ray images).
• Scalability - Since sets of medical images are
continuously increasing in size, we will build pre-
dictive models that can take advantage of an in-
cremental availability of training data.
4 METHODOLOGY
We make a distinction between two stages: (1) devel-
opment of novel predictive models for medical image
analysis, leveraging techniques for transfer learning
and unsupervised deep learning, and (2) an extensive
quantitative evaluation of the newly developed predic-
tive models.
4.1 Development
• Datasets - Starting from a mammography image
dataset for the purpose of detecting breast can-
cer, several additional medical image datasets will
be selected, related to different image modalities
(e.g., X-ray and Computed Tomography (CT))
and diseases (e.g., diabetic retinopathy, tubercu-
losis, and lung cancer). The selection of proper
datasets will be followed by data-specific prepro-
cessing.
• Source Network - As our source network, we will
make use of Inception V4 (Szegedy et al., 2016b),
a deep neural network architecture developed by
Google. We have selected this network because it
achieved the best top-5 accuracy in 2016 for the
task of image recognition (that is, a top-5 accu-
racy of 95.2%), outperforming other state-of-the-
art deep neural networks. Also, since it is a deep
neural network with repeated inception blocks, we
can easily observe the occurrence of overfitting,
and a poor effectiveness of prediction in general,
when doing vanilla training by means of a small
dataset. Thus, we will demonstrate the effective-
ness of our approach by comparing the results ob-
tained through vanilla training with the results ob-
tained through transfer learning and fine-tuning.
• Vanilla Training - We will train the source net-
work on a given dataset, using the network ob-
tained as a baseline.
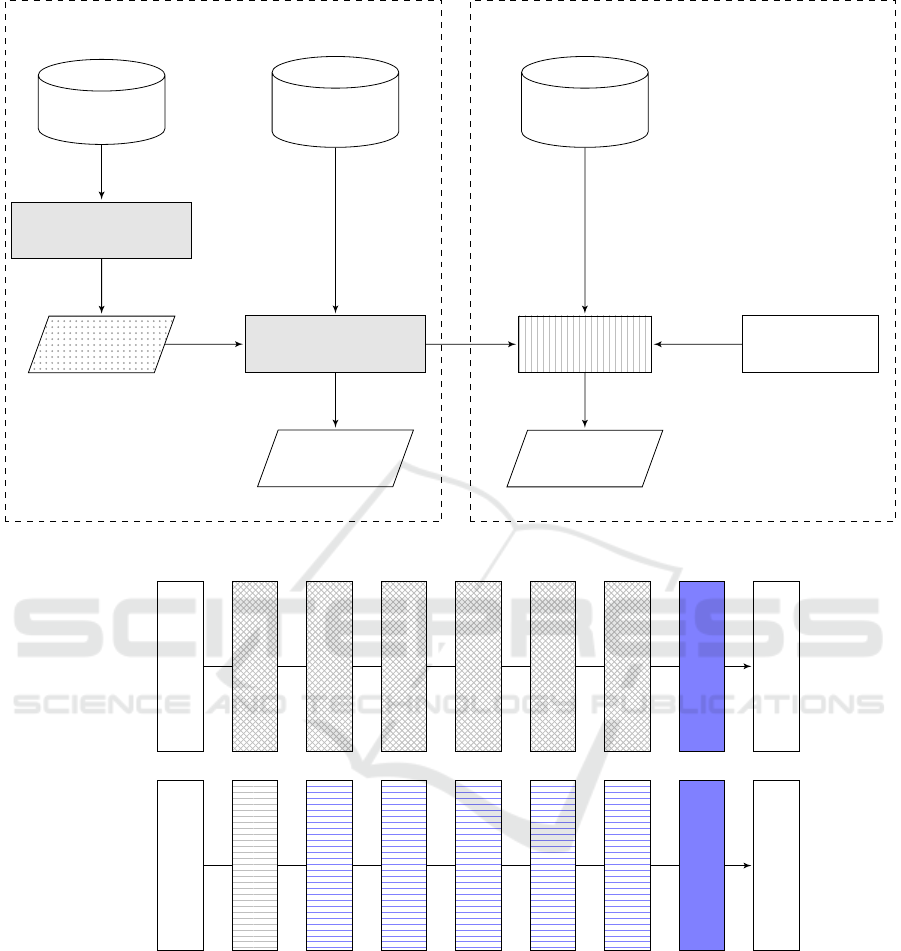
• Tranfer Learning with Fine-tuning - As shown
in Figure 2, transfer learning will be performed,
followed by fine-tuning. In our research, we will
experiment with different strategies for transfer
leaning and fine-tuning.
• Data Augmentation - Depending on the dataset
used, various techniques for data augmentation
will be implemented for the purpose of vanilla
training and transfer learning. Commonly used
data augmentation techniques are rotation, verti-
cal flipping, horizontal flipping, translation, con-
trast enhancement, and saturation.
• Unsupervised Learning - Considering the pres-
ence of unlabeled images and the data-hungry na-
ture of deep learning techniques, we will develop
unsupervised neural networks, combining VAEs
and DCGANs. For example, samples can be gen-
erated by a VAE, and these samples can then be
investigated by a deep discriminator, constructed
through transfer learning, so to see whether the
samples are real (representative) or fake (non-
representative) in nature.
4.2 Evaluation
Our research will primarily focus on assessing the ef-
fectiveness of the novel predictive models developed.
In practice, the effectiveness of deep learning-based
image classification is determined by calculating met-
rics like accuracy, recall, precision, and F-measure.
When it comes to medical imaging analysis, we need
to consider two additional metrics, namely specificity
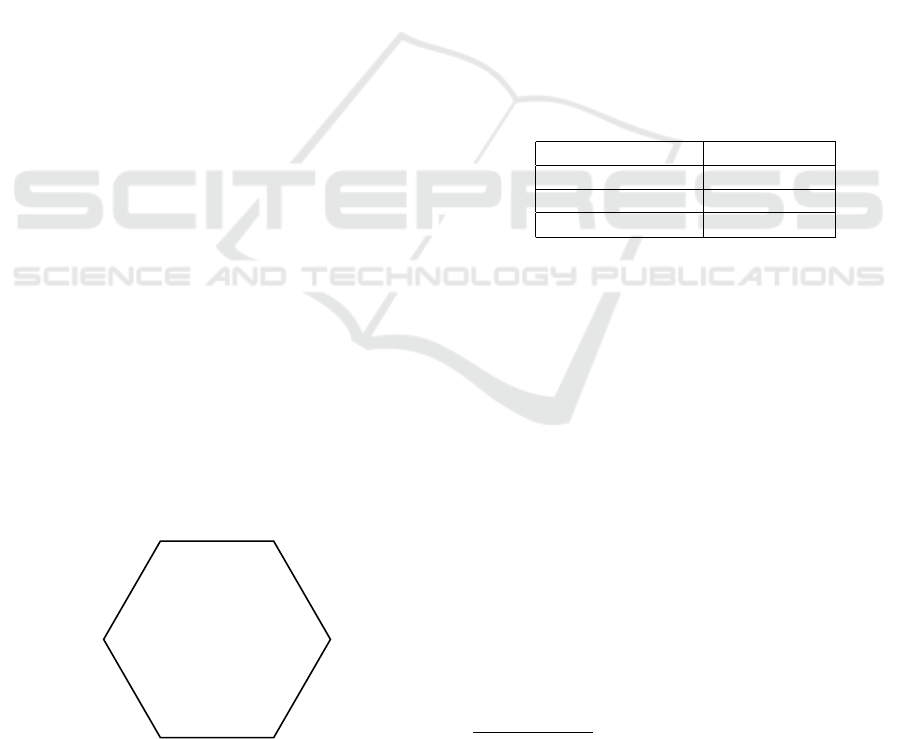
and ROC curve. Thus, as shown in Figure 3, we will
make use of the following six metrics in our doctoral
research:
• Accuracy - This is one of the most important met-
rics to evaluate the effectiveness of a predictive
model. It refers to the closeness between the com-
puted outcomes and the diagnosed labels.
• Recall - This metric, which is also known as sen-
sitivity, measures the proportion of positives that
are correctly identified as such.
• Precision - This metric indicates how closely the
computed outcomes are to the diagnosed labels,
regardless of the accuracy.
• F-measure - This metric is the harmonic mean of
recall and precision. When equally weighted, we
refer to this metric as F1. Depending on the pur-
pose of a particular research effort, we can place
more weight on recall (F2) or precision (F0.5).
• Specificity - This metric measures the proportion
of negatives that are correctly identified as such.
Together with recall, it is considered to be one of
the most important metrics in the area of medical
Towards Novel Methods for Effective Transfer Learning and Unsupervised Deep Learning for Medical Image Analysis
35
ImageNet
Medical
dataset 1
Deep neural network
Learned
features
Deep neural network
Diagnosis
Deep
discriminator
Medical
dataset 2
Deep generator
Diagnosis
Phase 1 Phase 2
Data aug-
mentation
Data aug-
mentation
Transfer
features
Fine-tuning
Transfer
features
Data aug-
mentation
Generated
sample
images
Fine-tuning
Input layer
A
Softmax
Output layer
Input layer
B
Softmax
Output layer
Figure 2: Overview of transfer learning. Top: In Phase 1, the network to the left, as visualized by means of gray boxes, has
been trained on ImageNet from scratch. The learned features are then transferred to another network that focuses on medical
image analysis. Since the two use cases come with different class sizes, the last layer is retrained through fine-tuning. The
same eventually holds true for one or more preceding layers. In Phase 2, the deep discriminator is trained using a second
medical dataset, leveraging transferred features through fine-tuning. Sample images produced by the deep generator will be
fed into the deep discriminator, and finally, the diagnosis will come out as a result. Bottom: The network is a simplified
version of Inception V4, coming with six inception blocks and one softmax layer right before the output. In the A model, the
crosshatched blocks remain frozen as explained above, and the sofmax layer that is retrained is the layer marked in blue. In
the B model, the blue horizontally lined blocks can be optionally retrained through fine-tuning.
image analysis (Pewsner et al., 2004; Weinstein
et al., 2005).
• ROC Curve - This metric represents the relation
between the true positive fraction and the false
positive fraction (Hajian-Tilaki, 2013).
The accuracy, the recall, the precision, and the F1
score will help in preventing our models from suffer-
ing from the accuracy paradox, whereas the recall, the
DCBIOSTEC 2017 - Doctoral Consortium on Biomedical Engineering Systems and Technologies
36
specificity, and the ROC curve will help in demon-
strating the validity of our models.
5 EXPECTED OUTCOME
In our doctoral research, we will develop new end-
to-end learning tools for the construction of novel
predictive models that target medical diagnosis (see
Phase 2 in Figure 2). The novel predictive models are
intended to be optimal in terms of (1) reliability, (2)
transferability, and (3) scalability.
6 STAGE OF THE RESEARCH
Thus far, we have performed the steps below, using
the mammography dataset discussed in Section 6.1:
1. Preprocessing of the dataset.
2. Application of different types of deep learning
techniques, either from scratch or by making use
of pre-training and transfer learning.
3. Evaluation of the effectiveness of the different
techniques using several metrics, namely accu-
racy, sensitivity, and specificity.
In the following section, we summarize our pre-
liminary results.
6.1 Use Case: Breast Cancer
We have chosen mammography-based diagnosis of
breast cancer as our first use case, relying on the pub-
licly available Digital Database for Screening Mam-
mography (DDSM) (Bowyer et al., 1996) (Heath
et al., 1998).
Breast cancer is the most commonly diagnosed
cancer among women. According to the U.S.
Recall
Accuracy
ROCcurve
Speci f icity
F − measure
Precision
Figure 3: A hexagon chart for plotting six metrics. This
hexagon chart will help in visualizing and comparing the
effectiveness of the newly developed predictive models.
Breast Cancer Statistics published in 2016 (BREAST-
CANCER.ORG, 2016), about 12% of women in the
U.S. will develop invasive breast cancer over the
course of their lifetime. Moreover, one out of thou-
sand men are also at risk of developing breast cancer.
A timely diagnosis of breast cancer can help in im-
proving the quality of life of a patient. However, mak-
ing a timely diagnosis is not easy, given that early-
stage lesions are difficult to detect in mammography
images. Moreover, human errors can lead to a faulty
diagnosis as well (Ertosun and Rubin, 2015).
The 10,412 images in the DDSM dataset were
originally formatted as Lossless JPEG (LJPEG). We
converted these images to the Portable Network
Graphics (PNG) format by means of a utility available
on (Sharma, 2015). The images in the DDSM dataset
can also be categorized into two types, depending on
the way acquisition was done: Cranial-Caudal (CC)
view images and MedioLateral-Oblique (MLO) view
images. Each type of image comes with a left- and
right-side version per patient, thus resulting in a total
of four images per patient.
Table 1: Information about DDSM.
Size 10,412
Type Mammogram
Format PNG
Positive:negative
1
4:6
6.2 Experiments
We performed a first experiment using the following
steps:
1. As illustrated by the leftmost image in Figure 4,
many mammogram images from the dataset used
contain a white border, black stains, text, and/or
noise. In addition, the size and the orientation of
the images may vary. Thus, to only feed regions-
of-interest to the network used, we preprocessed
the images, removing white borders, text, and
noise, followed by a resize operation.
2. As shown in Figure 4, a deep convolutional net-
work is trained by making use of the preprocessed
images. In this experiment, the network architec-
ture used was Inception V4. To measure the trans-
ferability of each model, (1) we trained models
from scratch and (2) we used a model pre-trained
on ImageNet. Data augmentation is used during
1
An image with a positive label indicates that a patient
definitely has one or more lesions that can be either benign
or malignant (gold standard in this case). On the other hand,
an image with a negative label means that the image under
consideration does not have any lesions.
Towards Novel Methods for Effective Transfer Learning and Unsupervised Deep Learning for Medical Image Analysis
37

Figure 4: Our overall research approach. In the input image to the left, both the text in the red circle and the white border are
removed so not to give unnecessary information to the predictive model used. In a next step, the input image is cropped and
resized so to ensure that all input images have the same dimension. The pre-processed input image is then fed to the predictive
model used for diagnosis purposes.
the training of each model. The data augmenta-
tion methods used in this experiment are vertical
flipping, horizontal flipping, enhancement of con-
trast, change in saturation, and random cropping.
3. We conducted a preliminary evaluation. The re-
sults obtained are discussed in the next section.
6.3 Results
Compared to the 95.2% of accuracy achieved by In-
ception V4 on the task of image recognition, the accu-
racy results shown in Table 2 are significantly lower.
Furthermore, and as expected, the experiments that
have been performed thus far suffered from overfit-
ting during both vanilla training and transfer learn-
ing, and where the latter was done by retraining the
last softmax layer. Nevertheless, the use of transfer
learning resulted in an accuracy and sensitivity that
is slightly higher than the accuracy and sensitivity of
vanilla training. Besides, the efficiency of transfer
learning was significantly higher than the efficiency
of vanilla training: two to three times, depending on
the number of layers retrained.
Table 2: Preliminary results obtained for the usage of In-
ception V4 as our baseline architecture. The asterisk indi-
cates that retraining of the underlying model started from
the last inception block, whereas retraining of the other
model started from the softmax layer.
Accuracy Sensitivity Specificity
Inception 69.80% 41.25% 88.43%
Inception* 72.00% 48.61% 87.94%
At the time of writing, further research using the
DDSM dataset is focusing on layer-wise fine-tuning
and on applying various combinations of different
data augmentation methods.
ACKNOWLEDGEMENT
The research effort described in this paper was funded
by Ghent University, the Ghent University Global
Campus, imec, Flanders Innovation & Entrepreneur-
ship (VLAIO), the Fund for Scientific Research-
Flanders (FWOFlanders), and the European Union.
REFERENCES
Ackley, D. H., Hinton, G. E., and Sejnowski, T. J. (1985). A
Learning Algorithm for Boltzmann Machines. Cogni-
tive Science, 9(1):147–169.
Azizpour, H., Sharif Razavian, A., Sullivan, J., Maki, A.,
and Carlsson, S. (2015). From Generic to Specific
Deep Representations for Visual Recognition. In Pro-
ceedings of CVPR.
Belghazi, I. (2016). Adversarially Learned Inference.
https://ishmaelbelghazi.github.io/ALI/.
Boesen, A., Larsen, L., Sønderby, S. K., Larochelle, H.,
and Winther, O. (2015). Autoencoding beyond Pixels
using a Learned Similarity Metric. In Proceedings of
ICML, pages 1558–1566.
Bowyer, K., Kopans, D., Kegelmeyer, W., Moore, R., Sal-
lam, M., Chang, K., and Woods, K. (1996). The
Digital Database for Screening Mammography. In
Third International Workshop on Digital Mammogra-
phy, volume 58, page 27.
Chen, X., Duan, Y., Houthooft, R., Schulman, J., Sutskever,
I., and Abbeel, P. (2016). InfoGAN: Interpretable
Representation Learning by Information Maximizing
Generative Adversarial Nets. In Proceedings of NIPS
2016.
Chen, X.-W. and Lin, X. (2014). Big Data Deep Learning:
Challenges and Perspectives. IEEE Access, 2:514–
525.
Dosovitskiy, A. and Brox, T. (2016). Generating Images
DCBIOSTEC 2017 - Doctoral Consortium on Biomedical Engineering Systems and Technologies
38

with Perceptual Similarity Metrics based on Deep
Networks. In arXiv preprint arXiv:1602.02644.
Ertosun, M. G. and Rubin, D. L. (2015). Probabilistic
Visual Search for Masses within Mammography Im-
ages using Deep Learning. In 2015 IEEE Interna-
tional Conference on Bioinformatics and Biomedicine
(BIBM), pages 1310–1315.
Frans, K. (2016). Generative Adversarial Networks
Explained. http://kvfrans.com/generative-adversial-
networks-explained/.
Girshick, R., Donahue, J., Darrell, T., and Malik, J. (2014).
Rich Feature Hierarchies for Accurate Object Detec-
tion and Semantic Segmentation. In Proceedings of
the IEEE Conference on Computer Vision and Pattern
Recognition, pages 580–587.
Goodfellow, I., Bengio, Y., and Courville, A. (2016). Deep
Learning. Book in preparation for MIT Press.
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B.,
Warde-Farley, D., Ozair, S., Courville, A., and Ben-
gio, Y. (2014). Generative Adversarial Nets. In
Advances in Neural Information Processing Systems,
pages 2672–2680.
Guttenberg, N., Sinapayen, L., Yu, Y., Virgo, N., and Kanai,
R. (2016). Recurrent Generative Auto-encoders and
Novelty Search. http://www.araya.org/archives/1306.
Hajian-Tilaki, K. (2013). Receiver Operating Characteris-
tic (ROC) Curve Analysis for Medical Diagnostic Test
Evaluation. Caspian Journal of Internal Medicine,
4(2):627.
Heath, M., Bowyer, K., Kopans, D., Kegelmeyer Jr,
P., Moore, R., Chang, K., and Munishkumaran, S.
(1998). Current Status of the Digital Database for
Screening Mammography. In Digital Mammography,
pages 457–460. Springer.
Hinton, G., Deng, L., Yu, D., Dahl, G. E., Mohamed, A.-
r., Jaitly, N., Senior, A., Vanhoucke, V., Nguyen, P.,
Sainath, T. N., et al. (2012). Deep Neural Networks
for Acoustic Modeling in Speech Recognition: The
Shared Views of Four Research Groups. IEEE Signal
Processing Magazine, 29(6):82–97.
Kingma, D. P. and Welling, M. (2013). Auto-encoding Vari-
ational Bayes. arXiv preprint arXiv:1312.6114.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012). Im-
agenet Classification with Deep Convolutional Neural
Networks. In Advances in Neural Information Pro-
cessing Systems, pages 1097–1105.
Lalkhen, A. G. and McCluskey, A. (2008). Clinical Tests:
Sensitivity and Specificity. Continuing Education in
Anaesthesia, Critical Care & Pain, 8(6):221–223.
LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep Learn-
ing. Nature, 521(7553):436–444.
Mikolov, T., Sutskever, I., Chen, K., Corrado, G. S.,
and Dean, J. (2013). Distributed Representations of
Words and Phrases and Their Compositionality. In
Advances in Neural Information Processing Systems,
pages 3111–3119.
Pan, S. J. and Yang, Q. (2010). A Survey on Transfer Learn-
ing. IEEE Transactions on Knowledge and Data En-
gineering, 22(10):1345–1359.
Pewsner, D., Battaglia, M., Minder, C., Marx, A., Bucher,
H. C., and Egger, M. (2004). Ruling a Diagnosis In
or Out with SpPIn and SnNOut: a Note of Caution.
BMJ, 329(7459):209–213.
Radford, A., Metz, L., and Chintala, S. (2015). Unsu-
pervised Representation Learning with Deep Convo-
lutional Generative Adversarial Networks. CoRR,
abs/1511.06434.
Rezende, D. J., Mohamed, S., and Wierstra, D. (2014).
Stochastic Backpropagation and Approximate Infer-
ence in Deep Generative Models. arXiv preprint
arXiv:1401.4082.
Rumelhart, D. E., Hinton, G. E., and Williams, R. J. (1985).
Learning Internal Representations by Error Propaga-
tion. Technical report, DTIC Document.
Salimans, T., Goodfellow, I., Zaremba, W., Cheung, V.,
Radford, A., and Chen, X. (2016). Improved Tech-
niques for Training GANs. In Proceedings of NIPS
2016.
Salimans, T., Kingma, D. P., Welling, M., et al. (2015).
Markov chain Monte Carlo and Variational Inference:
Bridging the Gap. In International Conference on Ma-
chine Learning, pages 1218–1226.
Santoro, A., Bartunov, S., Botvinick, M., Wierstra,
D., and Lillicrap, T. (2016). One-shot Learning
with Memory-Augmented Neural Networks. arXiv
preprint arXiv:1605.06065.
Sharma, A. (2015). DDSM Utility.
https://github.com/trane293/DDSMUtility.
Szegedy, C., Ioffe, S., and Vanhoucke, V. (2016a).
Inception-v4, Inception-ResNet and the Impact of
Residual Connections on Learning. arXiv preprint
arXiv:1602.07261.
Szegedy, C., Ioffe, S., and Vanhoucke, V. (2016b).
Inception-v4, Inception-ResNet and the Impact of
Residual Connections on Learning. arXiv preprint
arXiv:1602.07261.
BREASTCANCER.ORG (2016). U.S. Breast Cancer
Statistics. http://www.breastcancer.org/symptoms/
understand
bc/statistics.
Weinstein, S., Obuchowski, N. A., and Lieber, M. L. (2005).
Clinical Evaluation of Diagnostic Tests. American
Journal of Roentgenology, 184(1):14–19.
Wong, T. Y. and Bressler, N. M. (2016). Artifi-
cial Intelligence With Deep Learning Technology
Looks Into Diabetic Retinopathy Screening. JAMA,
316(22):2366–2367.
Yosinski, J., Clune, J., Bengio, Y., and Lipson, H. (2014).
How Transferable are Features in Deep Neural Net-
works? In Proceedings of NIPS, pages 3320–3328.
Towards Novel Methods for Effective Transfer Learning and Unsupervised Deep Learning for Medical Image Analysis
39
