
A Data-aware MultiWorkflow
Scheduler for Clusters on WorkflowSim
C
´
esar Acevedo, Porfidio Hern
´
andez, Antonio Espinosa and Victor M
´
endez
Computer Architecture and Operating System, Univ. Aut
´
onoma de Barcelona, Barcelona, Spain
Keywords:
MultiWorkflow, Data-aware, Cluster, Simulation, Storage Hierarchy.
Abstract:
Most scientific workflows are defined as Direct Acyclic Graphs. Despite DAGs are very expressive to re-
flect dependencies relationships, current approaches are not aware of the storage physiognomy in terms of
performance and capacity. Provide information about temporal storage allocation on data intensive applica-
tions helps to avoid performance issues. Nevertheless, we need to evaluate several combinations of data file
locations and application scheduling. Simulation is one of the most popular evaluation methods in scientific
workflow execution to develop new storage-aware scheduling techniques or improve existing ones, to test scal-
ability and repetitiveness. This paper presents a multiworkflow store-aware scheduler policy as an extension
of WorkflowSim, enabling its combination with other WorkflowSim scheduling policies and the possibility
of evaluating a wide range of storage and file allocation possibilities. This paper also presents a proof of
concept of a real world implementation of a storage-aware scheduler to validate the accuracy of the Work-
flowSim extension and the scalability of our scheduler technique. The evaluation on several environments
shows promising results up to 69% of makespan improvement on simulated large scale clusters with an error
of the WorflowSim extension between 0,9% and 3% comparing with the real infrastructure implementation.
1 INTRODUCTION
Direct Acyclic Graphs (DAGs) show an easy and
meaningful way of defining task dependence relation-
ships like those typically found in a scientific applica-
tion workflow. It is very common to exploit described
dependencies to apply job scheduling policies. De-
spite their wide usage to represent application stages,
it is not easy to infer any information on task data ac-
cess. That is, there is no information on how input,
output or intermediate data are accessed from CPU
once tasks are running.
In production infrastructures, a significant part of
the scientific computing workload is corresponding to
data intensive applications. This is turning the design
and setup of data-aware schedulers into a critical topic
to make a better use of the computing and storage
resources, as well as to improve the response times.
We may find data-aware scheduling approaches for
different purposes. For example, there are solutions
that consider distributed data locations such as (Zhang
et al., 2016) or frequent use of data files like (Del-
gado Peris et al., 2016) does. Our approach is focused
in a cluster environment without remote data distri-
bution. Our motivation is to provide a data-aware
logic able to deal with different storage performance
and space capacity, as well as to offer the possibility
of integrating such approach with any logical work-
flow scheduling in WorkflowSim(Chen and Deelman,
2012), which is one of the most widespread workflow
simulators both for theoretical scheduling design and
to real infrastructure setup.
When data files are going to be read several times,
we try to reduce communication time of data inten-
sive applications by locating files close to the com-
putation nodes in a specific storage level. Due to
the peculiarity of scientific workflows composed of
many applications sharing files, we can take advan-
tage of having file copies at multiple storage hierar-
chy levels such as distributed file system (NFS), Local
Hard Disk (HDD), Local Solid State Disk(SSD), Lo-
cal Ramdisk and Local Cache. We are making use of
this complete storage hierarchy in a proposed Shared
Input File Policy.
Bioinformatics applications are characteristic of
many scientific workflows, due to the data intensive
workload patterns and the variety of the workflow
structures. In the present work we focus on bioinfor-
matics workflows for testing purposes, because they
represent a wide range of scientific applications, par-
Acevedo, C., Hernández, P., Espinosa, A. and Mendez, V.
A Data-aware MultiWorkflow Scheduler for Clusters on WorkflowSim.
DOI: 10.5220/0006303500790086
In Proceedings of the 2nd International Conference on Complexity, Future Information Systems and Risk (COMPLEXIS 2017), pages 79-86
ISBN: 978-989-758-244-8
Copyright © 2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
79
ticularly in the scenarios of high level of complexity.
Hereby, bioinformatics applications performing
operations like short read mapping, sequence align-
ment and variant analysis usually work as batches.
Batches are multiworkflows with the same reference
or input data file to be read. This approach allows to
transform a dynamic problem into a static problem.
Presented implementation only considers groups
of workflows as batches to be scheduled. The data
file size and location needs to be taken into account
on the specifications of workflow abstract graph, to
exploit large hierarchical storage systems.
A representative trait of workflow structures found
in bioinformatics applications is the use of the same
data input for different executions, even different ap-
plications. Our scheduler exploits the storage hier-
archy to allocate most used data files. We present in
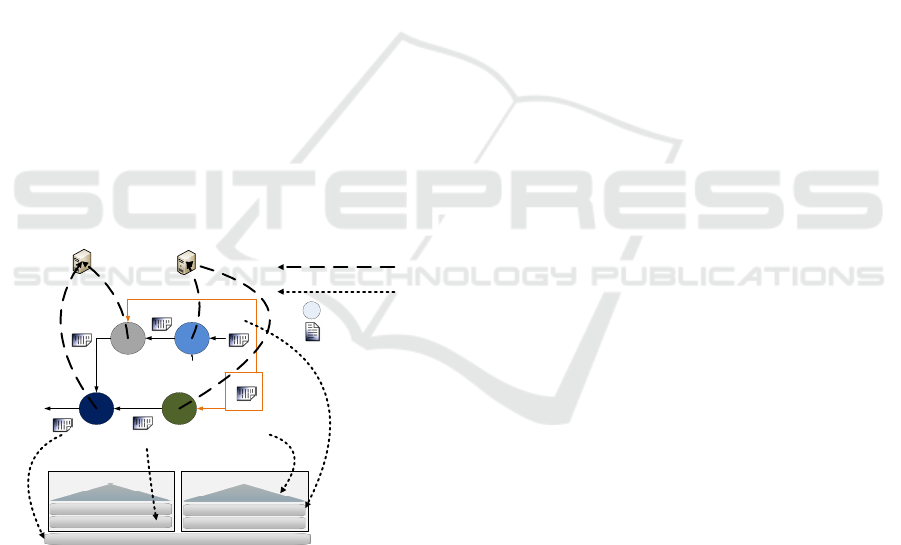
figure 1 a scheduler which needs to decide the best lo-
cation of input, temporal and output data files of one
workflow on a hierarchical storage system. As each
file can be located in any level, there are many pos-
sible combinations and our aim is to find out the best
option.
The scheduling target is to use this file location in-
formation to better assign application tasks to a com-
puting node. In our case, we use a storage hierarchy
with 4 levels: NFS and a Local Storage for each node
composed of Local RamDisk, SSD and HDD.
Reads
File
Temp
Temp
Temp
Reference
File
Result
Fast2Sanger
Gatk
BWA
SamTools
Application
Data file
Distributed File System
Node 1
Node 2
Local Storage
RAMDISK
SSD
HDD
Local Storage
RAMDISK
SSD
HDD
Scheduling Application
Scheduling Data File
Figure 1: Data and App location of workflow scheduling.
In a previous work (Acevedo et al., 2016) we in-
troduced an implementation of our data-aware mul-
tiworkflow policy considering Local HDD, Local
Ramdisk, Local SSD and NFS versus a classic List
Scheduling with NFS running in a prototype cluster.
The contribution of the present work is enhanc-
ing previous approach with a Shared Input File Pol-
icy, which contemplates an exhaustive analysis of all
the possible data file locations in any storage hierar-
chy level, together with any application assignment to
any CPU node. In this sense, this paper presents a so-
lution to alleviate the complexity of evaluating all the
storage and computing possibilities. For that, we re-
quire the use of a simulator to find which storage level
usage has more impact on data access latency, evalu-
ating in single workflow of particular application and
multiworkflow executions to be more realistic.
Our approach is first to provide an extension of
a simulator to develop new storage-aware schedulers
and second to validate proposed simulator extensions
with a list of experiments taken from our prototype
cluster. Then, use this simulator to evaluate the scala-
bility of our storage-aware proposal in larger compu-
tational environments.
To this end, we have started from WorkflowSim,
a multiworkflow simulator developed by (Chen and
Deelman, 2012) for grid and cluster environments
with build-in schedulers such as HEFT, Min-Min,
Max-Min, FCFS and Random. Nevertheless, there
was no storage-aware scheduler ready to be used in
the original simulator. To improve this situation, we
added the ability to exploit data locations by adding
storage hierarchy levels as Ramdisk and SSD and to
use them along with NFS and Local Hard Disk. Then,
we could begin the implementation of any state of the
art data-aware schedulers, we can improve them and
we can develop new ones.
We will compare our proposed data-aware sched-
uler against HEFT, Min-Min, Max-Min, FCFS and
Random schedulers on WorkflowSim for applica-
tions with shared input files with sizes of 2048Mb,
1024Mb, and 512Mb and up to 1024 cores.
Due to the multiple possibilities to locate data
files, the simulator gave us the ability to explore all
combinations prior to apply the best location policy
into real cluster storages. We used NFS as initial stor-
age and started locating reference and temporal files
in Local RamDisk, HDD or SSD. Then, we had up
to 69% of makespan improvement on simulated large
scale clusters with an error between 0,9% and 3%.
The rest of the paper is organized as follows. Re-
lated work is discussed in Section II. Then we de-
scribe how the scheduler is attached to the Work-
flowSim simulator in Section III. Section IV elabo-
rates the experiment design and evaluates the perfor-
mance of proposed algorithm. Finally, we summarize
and lay out the future work in Section V.
2 RELATED WORK
For list scheduling such as cited in (Ilavarasan
and Thambidurai, 2007),(Bolze et al., 2009) and
(Topcuoglu et al., 1999), there is not much research
COMPLEXIS 2017 - 2nd International Conference on Complexity, Future Information Systems and Risk
80

that considers that many applications have evolved
from compute-intensive to data-intensive in High Per-
formance Cluster environments. Neither workflow-
aware scheduling, as proposed by (Costa et al., 2015)
and (Vairavanathan et al., 2012) that provides meth-
ods to expose data location information, that gener-
ally are hidden, to exploit a per-file access optimiza-
tion. MapReduce (Dean and Ghemawat, 2008) is an
environment for data-intensive applications that lo-
cates data files to a specific data storage node prior
to execution but it is specific for certain patterns of
workflows. A DAG-based workflow scheduling tech-
nique for Condor environment has been studied by
(Shankar and DeWitt, 2007). In the field of Cloud
computing, (Bryk et al., 2016) shows a study of
storage-aware algorithm. For (Ramakrishnan et al.,
2007) research looks to improve overall performance
of scientific workflows by improving location over
distributed data storage with disk constraints. CoScan
(Wang et al., 2011) studies how caching input files im-
proves execution time when several applications are
going to read the same information.
Data location research has been done with exam-
ples like PACman (Ananthanarayanan et al., 2012),
RamCloud (Ousterhout et al., 2011), and RamDisk
(Wickberg and Carothers, 2012) that provides data lo-
cation techniques but not all of them in a cluster-based
environment. When dealing with scientific applica-
tions, input files are usually shared by many of them.
Then, our aim is to apply techniques to move data files
to a faster storage level that is closer to the compute
node.
Many scientific fields commonly repeat experi-
ments and reuse workflows and data files. In this
context, many users execute the same workflows or
applications daily. Due to this repetitiveness, we ex-
pand the problem from single workflow to multiwork-
flow. Research like (Barbosa and Monteiro, 2008)
uses list scheduling heuristics. For (Zhao and Sakel-
lariou, 2006) and (H
¨
onig and Schiffmann, 2006) a
meta-scheduler for multiple DAGs shows a way of
merging multiple workflows into one, to expose the
information about data location and improve the over-
all parallelism previous to a scheduling stage.
Nevertheless, there are multiple possible configu-
rations for scheduling applications and their data files
over the storage hierarchy. To reduce cost and time,
simulation has been proposed as a suitable system
for the evaluation of a range of workflow schedul-
ing strategies. In this way, we can find simulation
Manufacturing Agent Simulation Tool (MAST) (Mer-
dan et al., 2008) based on multi-agent negotiation,
where each resource agent performs local scheduling
using dispatching rules. Also, (Hirales-Carbajal et al.,
2010) proposes tGSF as a framework for scheduling
workflow on Grid. Both simulators were specifically
designed for analyzing a few aspects of workflow
scheduling considering infrastructures such as Grid.
CloudSim (Calheiros et al., 2011) is a framework that
models single workloads and simulates cloud com-
puting infrastructures. CloudSim lacks of the concept
of dependencies and does not consider overheads for
specific infrastructure configurations. WorkflowSim
(Chen and Deelman, 2012) extends CloudSim to im-
plement dependencies as workflows and allows us
to introduce the corresponding overhead to a cluster
storage level that is different than a cloud or grid ser-
vice. In any case, WorkflowSim lacks of a scheduler
that uses a storage-aware plug-in to simulate the use
of fast storage systems like RamDisk, SSD or any
other to manage application file I/O.
For our proposal, we selected a data-aware
scheduling implemented with an abstract meta-
workflow model. We implemented a storage-aware
extension to the simulator and fed it with information
about application computation times from the proto-
type cluster, a pattern recognition design to evalu-
ate data location for different sizes of shared input
files. Then, our objective was to reduce data access
latency accessing the provided files. Additionally, we
provided an extension of the storage class for differ-
ent storage devices such as Ramdisk and SSD. This
should give a solid background to develop and evalu-
ate new data-aware scheduling techniques.
3 WorkflowSim EXTENSION
Data-Aware Multiworkflow Pre-Scheduler is based
on locating shared input files in a storage hierarchy
in order to achieve lower latency access to read data
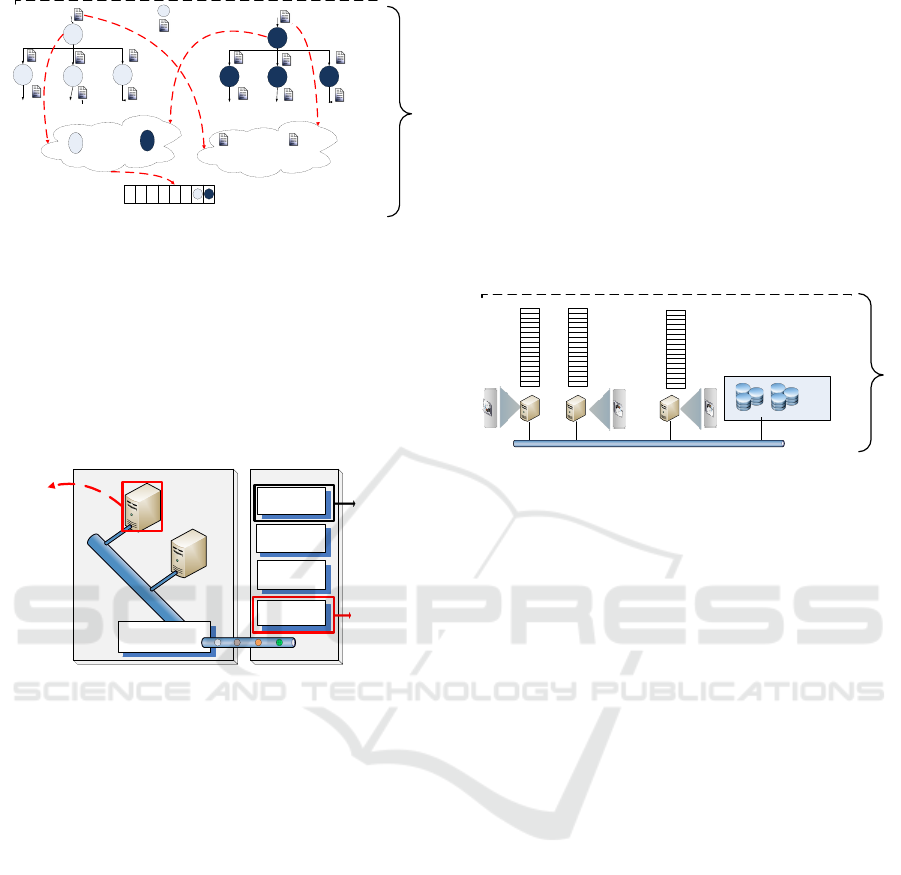
files. That is, according to figure 2 the pre-scheduling
step will accept a batch of applications from different
workflows. Then, it will merge all the applications
of different workflows into one meta-workflow with
dummy nodes at the beginning and end of the multi-
workflow. Next, it will apply a Critical Path analy-
sis to the resulting meta-workflow and locate files on
the hierarchical storage system according to parame-
ters like number and size of shared input files in the
scheduler level of the diagram. Finally, it will create a
priority list of applications to be sent to the scheduler.
We decided to use a well-known workflow sim-
ulator to improve and develop new storage-aware
scheduling techniques, compare with other sched-
ulers, and verify scalability of Data-Aware Multi-
workflow Cluster Scheduler on larger clusters. Work-
flowSim (Chen and Deelman, 2012) is used to simu-
A Data-aware MultiWorkflow Scheduler for Clusters on WorkflowSim
81
Application
Data File
INPUT
OUTPUT
OUTPUT
OUTPUT
Temp
Temp Temp
…..…...
……………..
……………..
Priority List
Pre Scheduling
INPUT
OUTPUT
OUTPUT
OUTPUT
Temp
Temp Temp
Figure 2: CPFL Pre-Scheduler design on Workflow Mapper
Module.
late workflows that have been modeled by DAGs de-
fined through XML files and implements various clas-
sic scheduling algorithms like HEFT, Min-Min, Max-
Min, FCFS and Random. In figure 3 we present the
main modules of the simulator. We highlight those
modules extended to implement our proposal.
EXECUTION
SITE
Ramdisk
SSD
LocalDisk
NFS
SUBMIT HOST
WORFLOW
MAPPER
WORFLOW
ENGINE
CLUSTERING
ENGINE
WORFLOW
SCHEDULER
Data-Aware
Scheduling
MULTI
WF
Crithical Path Queue
WORKER
NODE
REMOTE
SCHEDULER
WORKER
NODE
Figure 3: WorkFlowSim components (Chen and Deelman,
2012).
The workflow mapper is responsible of importing
several DAGs which are concatenated for multiwork-
flow execution. Then, it creates a list of applications
to be assigned to available resources. In any case, we
must enforce applications original dependencies to re-
spect the workflow natural execution order of prede-
cessors.
We introduced the Data-Aware scheduler in the
workflow mapper component following the guide
for extensions of WorkflowSim. We added a Java
CPFLScheduler file implementing the CPFL (Critical
Path File Location) data-aware scheduler described
above under org.workflowsim.scheduling. This is
where the pre-scheduling layer is implemented ex-
tending the BaseSchedulingAlgorithm code.
When needed, the clustering engine is responsible
of encapsulating multiple applications within a single
job. A workflow scheduler, according to user-defined
criteria, effectively adds every job or application to a
queue of ready applications to be assigned to worker
nodes.
Our next modification is the addition of the stor-
age hierarchy to the worker node on the execution site
of the simulator. Figure 4 shows how the applica-
tion has been queued according to critical path per-
formed on the pre-scheduling layer. Applications are
assigned to those worker nodes where data files were
stored. Initially the data files are in the Distributed
File System and moved or copied to local storage such
as Hard Disk, Ramdisk or SSD. Files are located ac-
cording to their size and a how many times are they
requested. The pre-scheduling stage is in charge of
analyzing the workflow pattern to extract the number
of requests per file.
Distributed File System
……………..
Node 1 Node 2 Node N
Application Queue
Application Queue
Application Queue
RAMDISK
RAMDISK
RAMDISK
Scheduler
Figure 4: CPFL Scheduler design on Workflow Scheduler
Module.
We implemented new storage hierarchy system el-
ements under org.cloudbus.cloudsim. Namely, Ss-
dDriveStorage and RamdiskDriveStorage. Accord-
ingly, we introduced needed parameters to model the
latency, average seek time and maximum transfer rate
to the specific storage device according to product
specifications of the industry, as an infrastructure of
the system for the Scheduler Layer in figure 4. We
have implemented Ramdisk over a RAM DDR3-1333
SDRAM as local storage on the execution site, with a
fixed size of 6.2GB, max transfer rate of 1.8GB/s, la-
tency of 0.003ms and 0.001ms of average seek time.
For SSD Intel SSD 510 Series like, we created a fixed
size of 1TB and max transfer rate of 500MB/s, latency
of 0.06ms and average seek time of 0.1ms.
Finally, in order to execute the scheduler we must
define which storage elements are we going to use and
how many of them. This is how the step from pre-
scheduling in figure 2 merges workflows into a meta-
workflow and propose a priority list to be executed.
Data files in this step are located in the storage hier-
archy according to size and read factor. Scheduling
is shown in figure 4 where applications are assigned
to nodes where data files are already located to be ap-
plied in the execution site of the simulator, as seen in
figure 3.
To have the simulator working we use CPFL as
the main scheduler. As CPFL is a scheduler for clus-
ter systems, we define network latency to 0ms and
call the functions that create the storage hierarchy and
store defined disks in a list. Once we have the ID of
COMPLEXIS 2017 - 2nd International Conference on Complexity, Future Information Systems and Risk
82
a storage element and the list of data files with their
preferred location we calculate the time of reading the
data files considering the storage type latency.
Our objective is to take advantage of data locality
and shared input file characteristic of bioinformatics
multiworkflows to reduce the access time to disks in
the storage hierarchy.
We modified the Execution Site module of
WorkFlowSim, specifically the representation of the
worker nodes. Infrastructure parameters like number
of processors, RAM capacity, local storage capacity,
types of local storage are defined at worker node. We
needed to add a new storage type, the ramdisk, to
evaluate our proposal. Once we had the new storage
type, a new cluster was created with a given amount of
workers, with defined parameters like operating sys-
tem and latency costs between the storage hierarchy
levels.
4 EXPERIMENT DESIGN AND
EVALUATION
In this section, we introduce an experimental de-
sign to evaluate the extension of WorkflowSim us-
ing the CPFL MultiWorkflow Data-Aware Scheduler.
First, We comparatively evaluate our approach against
HEFT classic list scheduling on a distributed file sys-
tem (NFS) of a prototype local cluster in order to feed
the simulator with real results of scheduling with stor-
age hierarchy usage.
The cluster environment has 32 nodes, and each
of the nodes has a CPU with 4 cores at 2.0Ghz, 12GB
of main memory, and a local Ramdisk of 6.2GB. The
file systems that we are using are NFS as distributed
file system, an EXT4 local disk format and a TMPFS
Ramdisk.
In the described experiment platform, a large
amount of data analysis is done by running bioin-
formatics applications. Most workflow work is ap-
plied to different bioinformatics data analysis steps
as genome alignment, variant analysis, and common
data file format transformations.
We selected a list of commonly used well charac-
terized bioinformatics applications to test the work-
flow management system. Then, analyzed a repos-
itory of historical execution times, as shown in ta-
ble 1, to extrapolate relevant execution times and re-
source usage. These are the elements that compose
the synthetic workflows for our experimentation fol-
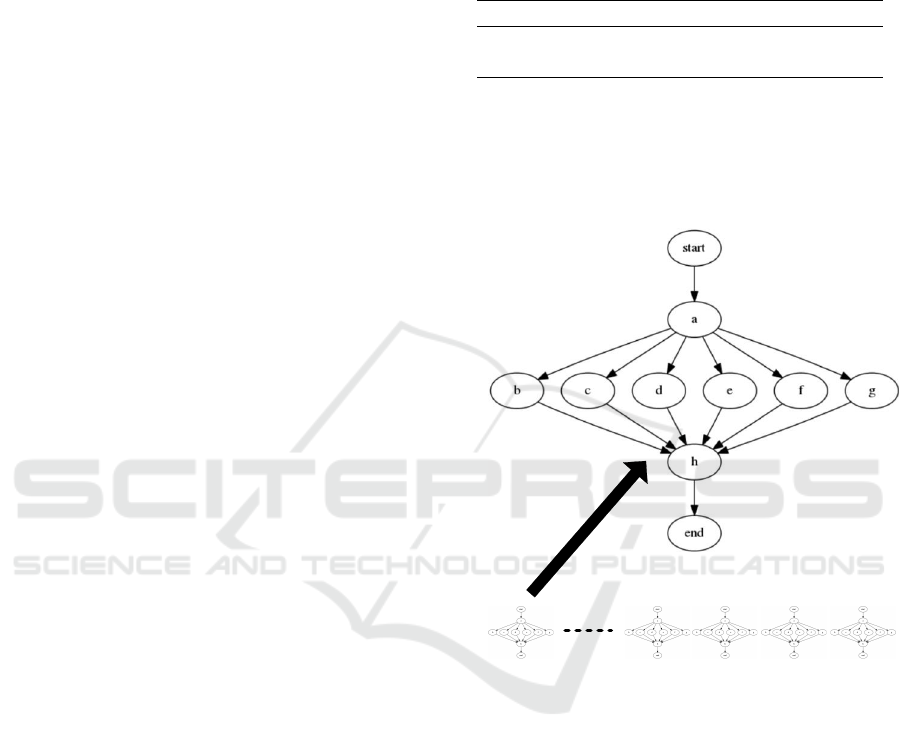
lowing the pattern shown in figure 1. BWA is an read-
mapper with CPU bound, Fast2Sanger and Sam2Bam
both are format transformers with I/O bound and Gatk
is a variant analyzer with CPU bound. Considering
the pattern of bioinformatics workflows, these appli-
cations and their dependencies as inputs, we have de-
signed a synthetic workflow pattern that we will use
for our experiments as we can see in figure 5.
Table 1: Workflow applications considered.
Apps (Syn Name) Exec IO IO RSS(Mb) CPU
Time(s) Read(Mb) Write(Mb) Util(%)
BWA (b,c,d) 11400 197 304 800 45
Fast2Sanger (a) 1440 67 69 180 98
Sam2Bam (h) 1020 160 54 480 99
Gatk (e,f,g) 1380 10 47 300 99
We show in figure 5, a graph schema correspond-
ing to the described workflows. A set of synthetic
multiworkflows based on the type of bioinformatics
data analysis done in the cluster with a batch of N
workflows in the system.
N-times
Figure 5: Synthetic Workflow Pattern.
We analyzed the workflow execution makespan
times as the main result of the policies in a previ-
ous study using a real cluster defined above. We ex-
ecuted 50 synthetic workflows with different shared
input files sizes: 512 MB, 1024 MB and 2048 MB
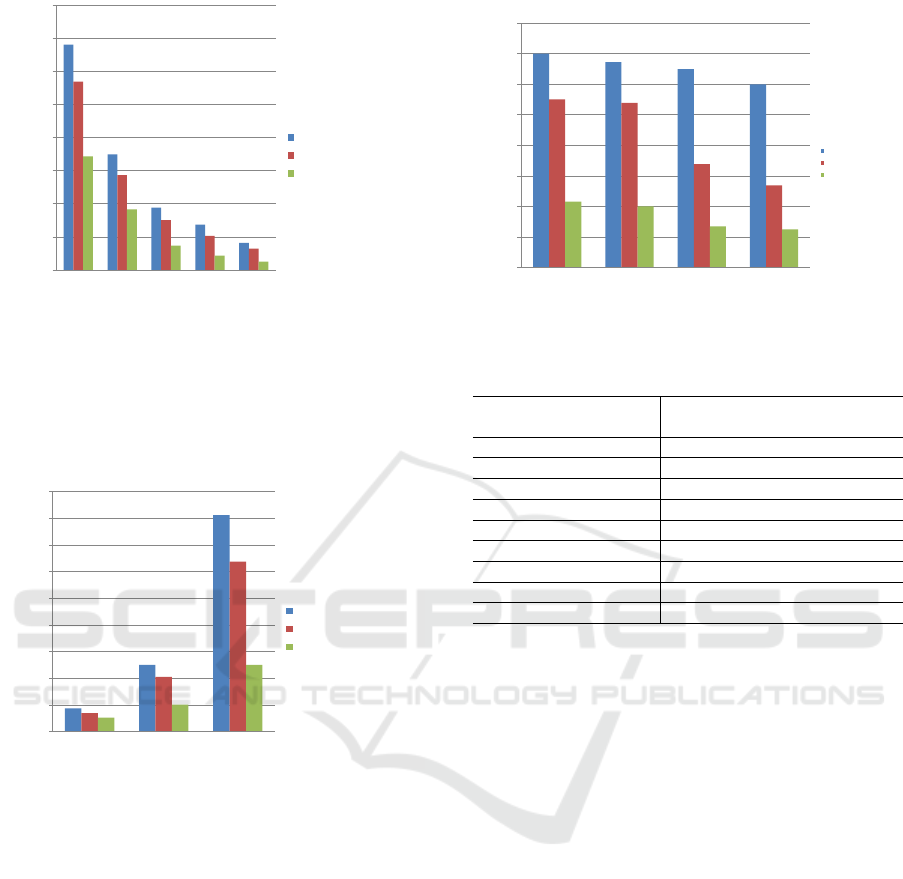
and found that the results were similar. In figure 6,
we show the comparative results for 2048MB. Con-
sidering the use of HEFT algorithm on a shared file
system (NFS) using between 8 and 128 cores, CPFL
obtained up to 50% better makespan when 2 storage
levels were used (local disk + Ramdisk). When only
one storage level was used (local disk), CPFL was
15% faster.
In figure 7 the results were similar considering
8, 16, 32, 64 and 128 cores. We present the results
for 128 cores where the gain of makespan was up to
70% when shared input files was 2048 MB and 40%
for 512 MB using 2 storage levels(Ramdisk + Local
A Data-aware MultiWorkflow Scheduler for Clusters on WorkflowSim
83
00
1728
3456
5184
6912
8640
10368
12096
13824
8 16 32 64 128
Seconds
Cores
HEFT (NFS)
CPFL (Local Disk)
CPFL (Local Disk + RamDisk)
Figure 6: Synthetic Workflow makespan with 2048MB of
shared input files.
Disk). Also, we show gains of 20% for 2048 MB files
and 12% for files of 512 MB when we used just local
disk.
00
173
346
518
691
864
1037
1210
1382
1555
512 1024 2048
seconds
Shared Input File Size (Mb)
128 Cores
HEFT (NFS)
CPFL (Local Disk)
CPFL (Local Disk + RamDisk)
Figure 7: Synthetic Workflow makespan on 128 cores.
Finally in figure 8 we can appreciate how reading
the same data files many times affects the makespan
of the multiworkflow execution. For 128 cores and
data files size of 2048MB we have a gain of up to 79%
for 16 shared files on CPFL approach versus a HEFT
on NFS storage. Nevertheless, we need to validate the
scalability of the shared input file beyond this point
when the system becomes stressed due to bigger data
files and multiworkflow batches size.
The next step is to use WorkflowSim to evaluate
how CPFL scales on larger clusters and to compare
with other heuristics.
WorkflowSim was tuned to behave as close as
possible to our local prototype cluster (IBM-like) in
which all simulation is executed. Compared in table
2 we can appreciate the differences with the standard
simulator specs without the extensions:
To evaluate the scalability we present results of
executing on a simulated cluster of 128, 256, 512 and
0
200
400
600
800
1000
1200
1400
1600
2 4 8 16
Seconds
Number of Shared Files
SIP 128 Cores - 2048MB
HEFT (NFS)
CPFL (Local Disk)
CPFL (Local Disk + RamDisk)
Figure 8: Synthetic workflow makespan on 128 cores with
many 2048MB Shared Input Files.
Table 2: Cluster Simulation Specs.
Specs Standard Test
Sim Sim (IBM-like)
Nodes p/Cluster 4 32
Processors p/ Node 1 4
Processors Freq. 1000 MIPS 6000 MIPS
RAM p/ Node 2 GB 12 GB
Disk Capacity p/ Node 1 TB 10 TB
SSD Capacity p/ Node - 1 TB
Ramdisk - 6.2 GB
Net Latency 0.2 ms 0 ms
Internal Latency 0.05 ms 0.05 ms
1024 cores. For this comparison, we use state of art
heuristics such as HEFT, MaxMin, MinMin, Random
and FCFS.
In figure 9, we present current 50 synthetic work-
flow execution on WorkflowSim. We obtained a
gain of 3.4% when we compare the use HEFT and
CPFL on NFS on 64 cores from 29622 to 30680
seconds; 5% better than MINMIN from 31188 sec-
onds; 4% better than MAXMIN from 31080 sec-
onds; 8% regarding FCFS from 32190 seconds and
21% respecting Random. When using CPFL on local
disk + Ramdisk + Local SSD, makespan has been re-
duced to 4134 seconds. Then, obtained gain is 54%
against HEFT, 54% against MINMIN, 55% regarding
MAXMIN a 56% respecting FCFS and up to 62% re-
specting to Random.
5 CONCLUSIONS AND OPEN
LINES
We extended WorkflowSim with storage hierarchy
levels as a way to help researchers to develop and im-
prove new storage-aware schedulers.
We modeled synthetic bioinformatics applications
COMPLEXIS 2017 - 2nd International Conference on Complexity, Future Information Systems and Risk
84

0
5000
10000
15000
20000
25000
30000
35000
40000
64 128 256 512 1024
Seconds
Cores
Synthetic
Ramdom
FCFS Sim - nfs
MINMIN Sim - nfs
MAXMIN Sim - nfs
HEFT Sim - nfs
CPFL Sim - nfs
CPFL Sim - ramd+disk+ssd
Figure 9: Synthetic CPFL simulation scaling to 1024 cores
versus others scheduling algorithms.
where some of them were sharing the same data files
as input. Techniques like caching shared input files
are desirable to prevent multiple file reads and to im-
prove the performance of the system I/O.
We used NFS as initial storage, locating reference
files and temporal files to Local RamDisk or Local
HDD or Local SSD obtained up to 69% of makespan
improvement on simulated large scale clusters with an
error between 0,9% and 3%.
Simulation of Synthetic Workflow Applications
has been correctly executed over a simulated cluster
tuned to behave like a real IBM cluster.
Even when a synthetic workflow has been used to
test scalability, our extension is able to simulate other
type of workflows due to the new storage hierarchy
added to WorkflowSim and the storage-aware sched-
uler.
As future work we are considering a list of op-
tions for data replacement polices in ramdisk, local
disk and SDD to further increase the efficiency of the
policies.
Looking forward, we plan to integrate this simula-
tor into a large cluster-based scientific workflow man-
ager like Galaxy (Goecks et al., 2010) which is a well
known workflow management system in the bioinfor-
matics community.
ACKNOWLEDGEMENTS
This work has been supported by project number
TIN2014-53234-C2-1-R of Spanish Ministerio de
Ciencia y Tecnolog
´
ıa (MICINN). This work is co-
founded by the EGI-Engage project (Horizon 2020)
under Grant number 654142.
REFERENCES
Acevedo, C., Hernandez, P., Espinosa, A., and Mendez, V.
(2016). A data-aware multiworkflow cluster sched-
uler. In Proceedings of the 1st International Confer-
ence on Complex Information Systems, pages 95–102.
SCITEPRESS.
Ananthanarayanan, G., Ghodsi, A., Wang, A., Borthakur,
D., Kandula, S., Shenker, S., and Stoica, I. (2012).
Pacman: coordinated memory caching for parallel
jobs. In Proceedings of the 9th USENIX conference
on Networked Systems Design and Implementation,
pages 20–20. USENIX Association.
Barbosa, J. and Monteiro, A. (2008). A list scheduling algo-
rithm for scheduling multi-user jobs on clusters. High
Performance Computing for Computational Science-
VECPAR 2008, pages 123—-136.
Bolze, R., Desprez, F., and Isnard, B. (2009). Evaluation
of Online Multi-Workflow Heuristics based on List-
Scheduling Algorithms. Gwendia report L.
Bryk, P., Malawski, M., Juve, G., and Deelman, E. (2016).
Storage-aware algorithms for scheduling of workflow
ensembles in clouds. Journal of Grid Computing,
14(2):359–378.
Calheiros, R. N., Ranjan, R., Beloglazov, A., De Rose,
C. A., and Buyya, R. (2011). Cloudsim: a toolkit for
modeling and simulation of cloud computing environ-
ments and evaluation of resource provisioning algo-
rithms. Software: Practice and Experience, 41(1):23–
50.
Chen, W. and Deelman, E. (2012). WorkflowSim: A toolkit
for simulating scientific workflows in distributed envi-
ronments. In 2012 IEEE 8th International Conference
on E-Science, e-Science 2012.
Costa, L. B., Yang, H., Vairavanathan, E., Barros, A., Ma-
heshwari, K., Fedak, G., Katz, D., Wilde, M., Ri-
peanu, M., and Al-Kiswany, S. (2015). The case for
workflow-aware storage: An opportunity study. Jour-
nal of Grid Computing, 13(1):95–113.
Dean, J. and Ghemawat, S. (2008). MapReduce: simplified
data processing on large clusters. Communications of
the ACM, 51(1):107–113.
Delgado Peris, A., Hern
´
andez, J. M., and Huedo, E. (2016).
Distributed late-binding scheduling and cooperative
data caching. Journal of Grid Computing, pages 1–
22.
Goecks, J., Nekrutenko, A., Taylor, J., and Team, T. G.
(2010). Galaxy : a comprehensive approach for sup-
porting accessible , reproducible , and transparent
computational research in the life sciences. Genome
biology.
Hirales-Carbajal, A., Tchernykh, A., R
¨
oblitz, T., and
Yahyapour, R. (2010). A grid simulation framework
to study advance scheduling strategies for complex
workflow applications. In Parallel & Distributed Pro-
cessing, Workshops and Phd Forum (IPDPSW), 2010
IEEE International Symposium on, pages 1–8. IEEE.
H
¨
onig, U. and Schiffmann, W. (2006). A meta-algorithm
for scheduling multiple dags in homogeneous sys-
tem environments. In Proceedings of the eighteenth
IASTED International Conference on Parallel and
Distributed Computing and Systems (PDCS’06).
A Data-aware MultiWorkflow Scheduler for Clusters on WorkflowSim
85

Ilavarasan, E. and Thambidurai, P. (2007). Low complexity
performance effective task scheduling algorithm for
heterogeneous computing environments. Journal of
Computer sciences.
Merdan, M., Moser, T., Wahyudin, D., Biffl, S., and Vrba,
P. (2008). Simulation of workflow scheduling strate-
gies using the mast test management system. In Con-
trol, Automation, Robotics and Vision, 2008. ICARCV
2008. 10th International Conference on, pages 1172–
1177. IEEE.
Ousterhout, J., Agrawal, P., Erickson, D., Kozyrakis, C.,
Leverich, J., Mazi
`
eres, D., Mitra, S., Narayanan, A.,
Ongaro, D., Parulkar, G., et al. (2011). The case for
ramcloud. Communications of the ACM, 54(7):121–
130.
Ramakrishnan, A., Singh, G., Zhao, H., Deelman, E.,
Sakellariou, R., Vahi, K., Blackburn, K., Meyers, D.,
and Samidi, M. (2007). Scheduling data-intensive
workflows onto storage-constrained distributed re-
sources. In Seventh IEEE International Symposium on
Cluster Computing and the Grid (CCGrid’07), pages
401–409. IEEE.
Shankar, S. and DeWitt, D. J. (2007). Data driven work-
flow planning in cluster management systems. In Pro-
ceedings of the 16th international symposium on High
performance distributed computing, pages 127–136.
ACM.
Topcuoglu, H., Hariri, S., and Wu, M.-Y. (1999).
Task scheduling algorithms for heterogeneous pro-
cessors. In Heterogeneous Computing Workshop,
1999.(HCW’99) Proceedings. Eighth, pages 3–14.
IEEE.
Vairavanathan, E., Al-Kiswany, S., Costa, L. B., Zhang,
Z., Katz, D. S., Wilde, M., and Ripeanu, M. (2012).
A workflow-aware storage system: An opportunity
study. In Proceedings of the 2012 12th IEEE/ACM
International Symposium on Cluster, Cloud and Grid
Computing (ccgrid 2012), pages 326–334. IEEE
Computer Society.
Wang, X., Olston, C., Sarma, A. D., and Burns, R. (2011).
Coscan: cooperative scan sharing in the cloud. In Pro-
ceedings of the 2nd ACM Symposium on Cloud Com-
puting, page 11. ACM.
Wickberg, T. and Carothers, C. (2012). The ramdisk storage
accelerator: a method of accelerating i/o performance
on hpc systems using ramdisks. In Proceedings of the
2nd International Workshop on Runtime and Operat-
ing Systems for Supercomputers, page 5. ACM.
Zhang, Y.-F., Tian, Y.-C., Fidge, C., and Kelly, W. (2016).
Data-aware task scheduling for all-to-all comparison
problems in heterogeneous distributed systems. Jour-
nal of Parallel and Distributed Computing, 93–94:87
– 101.
Zhao, H. and Sakellariou, R. (2006). Scheduling multi-
ple dags onto heterogeneous systems. In Proceedings
20th IEEE International Parallel & Distributed Pro-
cessing Symposium, pages 14–pp. IEEE.
COMPLEXIS 2017 - 2nd International Conference on Complexity, Future Information Systems and Risk
86
