
Localization of Demyelinating Plaques in MRI using Convolutional
Neural Networks
Bartłomiej Stasiak
1
, Paweł Tarasiuk
1
, Izabela Michalska
2
, Arkadiusz Tomczyk
1
and Piotr S. Szczepaniak
1
1
Institute of Information Technology, Lodz University of Technology, Wolczanska 215, 90-924 Lodz, Poland
2
Department of Radiology, Barlicki University Hospital, Kopcinskiego 22, 91-153 Lodz, Poland
Keywords:
Multiple Sclerosis, Convolutional Neural Networks.
Abstract:
In the paper a method of demyelinating plaques localization in head MRI sequences is presented. For that
purpose a convolutional neural network is used. It is trained to act as non-linear filter, which should indicate
(give a high response) in those image areas where the sought objects are located. Consequently, the output
of the proposed architecture is an image and not a single label as it is in the case of traditional networks with
pooling and fully connected layers. Another interesting feature of the proposed solution is the ability to select
network parameters using smaller patches cut from training images which reduces the amount of data that
must be propagated through the network. It should be emphasized that the conducted research was possible
only thanks to the manually outlined plaques provided by radiologist.
1 INTRODUCTION
In recent years convolutional neural networks (CNN)
allowed to make a significant progress in automatic
analysis of the images. It was possible thanks to the
technological progress (computations with GPU) and
access to large amount of labeled data. Labeled im-
age data, however, can be of different form. The most
popular (the easiest to gather) are data where the im-
age is accompanied by the label describing its con-
tent. This allows to train CNN solving a typical clas-
sification task. Other tasks, like precise localization
of objects (segmentation), require much more effort
to collect proper data. This task becomes even harder
if correct labeling of image content requires special-
ized, e.g. medical, knowledge. That is the case,
which is considered in this work where demyelinating
plaques are searched for within MRI (magnetic res-
onance imaging) data. The conducted research was
possible only thanks to the hard work of radiologist
who precisely outlined the regions of interest on ev-
ery slice of head MRI sequence.
As it was mentioned above, applying CNN to seg-
mentation task is not as popular as its application
to classification problems. Two basic groups of ap-
proaches can be found in the literature. First one is
a patch based approach where labels are assigned not
to the whole image but to the selected regions of that
image (in particular to the regions representing neigh-
bourhood of a given pixel). In other words it is a mod-
ified sliding window technique with CNN as a classi-
fier. This classifier, naturally, is not trained using the
whole image as an input. Instead, we use patches cut
from the training images, manually segmented by an
expert. Such a method was used, for example, in seg-
mentation of anatomical regions in MRI images (de
Brebisson and Montana, 2015). The second approach
uses a so-called fully convolutional approach (Shel-
hamer et al., 2016). In this case the whole image is
given as an input and as an output the image of the
same size, representing segmentation mask, is pro-
duced. To achieve such a functionality the network
has a special architecture. First some traditional con-
volutional and pooling layers are used, which reduces
the size of the resulting feature maps, and then some
upscaling (deconvolutional) layers are added to en-
large and combine those maps to obtain the image
of proper size. Such a fully convolutional network is
trained using whole images without the need of cut-
ting it into patches. This kind of approach was suc-
cessfully used in e.g. analysis of transmitted light
microscopy images (Milletari et al., 2016) and MRI
prostate examinations (Ronneberger et al., 2015). The
latter approach is particularly interesting since it con-
siders 3D convolution and the 3D MRI sequence is
processed by CNN as a whole.
Stasiak, B., Tarasiuk, P., Michalska, I., Tomczyk, A. and Szczepaniak, P.
Localization of Demyelinating Plaques in MRI using Convolutional Neural Networks.
DOI: 10.5220/0006298200550064
In Proceedings of the 10th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2017) - Volume 2: BIOIMAGING, pages 55-64
ISBN: 978-989-758-215-8
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
55

The solution proposed in this work to some extent
possesses features of both those approaches. On one
hand, it tries to train CNN to act as a non-linear filter
capable of indicating areas of interest. Consequently
the output is the image of the same size as the input.
In this case, however, no pooling is used and conse-
quently no upscaling is required. On the other hand, it
allows to train such a network using smaller patches
without the necessity of processing as large amount
of data as needed for the training based on the whole
images.
The paper is organized as follows: the second
section describes the considered dataset and medical
background justifying the importance of demyelinat-
ing plaques localization, in the third section the pro-
posed method is discussed and in the fourth and the
fifth section the obtained results and their analysis are
presented. Finally, the last section contains a short
summary of the conducted research.
2 MEDICAL BACKGROUND
Multiple sclerosis (MS) is a chronic autoimmune dis-
ease that attacks central nervous system and conse-
quently leads to neurological disability. The body’s
immune system destroys the nerve’s myelin sheaths
which form white matter in the brain and spinal cord.
The areas where the layer of myelin was damaged are
called demyelinating plaques and the whole is known
as demyelination. The diagnosis of the disorder is
made by the combination of clinical findings, the ex-
amination of cerebrospinal fluid and MRI of the cen-
tral nervous system. In patients with clinical symp-
toms suggesting MS the brain MR imaging can show
multifocal white matter lesions which are plaques of
demyelination. Nevertheless the process of demyeli-
nation can be a part of many other disorders, it is not
specific only to MS. The diagnosis of MS is more
likely if the plaques are distributed in some typical
areas in the brain such as: around the lateral ventri-
cles (periventricular), especially while they are orien-
tated perpendicularly to the long axis of the ventricles,
in the corpus callosum, along the boundary between
the white matter and cortex, in the cerebral and cere-
bellar peduncles, pons and medulla oblongata. The
most useful MRI scans for identifying white matter
lesions are T2-weighted images (T2WI), particularly
FLAIR sequences (fluid-attenuated inversion recov-
ery). On those images the demyelinating areas have
an abnormally high signal in comparison to the nor-
mal white matter. On T2WI both cerebrospinal fluid
and white matter lesions present a high signal so the
contrast between them is rather poor. In FLAIR tech-
nique the signal of cerebrospinal fluid is suppressed
what improves detecting of the white matter lesions,
especially in the periventricular distribution.
The present study has focused on marking the
lesions of demyelination on MR scans of the brain
(FLAIR sequences in axial plane). All magnetic reso-
nance images were obtained using 1,5 Tesla scanner.
The thickness of slices of each examination amounted
from 3mm to 5mm. The patient population consisted
of hundred people (fifty men and fifty women) of dif-
ferent age groups (between 19 and 66 years old). The
study has taken into consideration only patients with
confirmed diagnosis of MS. The severity of the dis-
ease differed from newly diagnosed to longstanding
disorder. The plaques of demyelination on magnetic
resonance images were defined as notable alterations
of the signal in the whole area occupied by the white
matter.
3 METHOD
Convolutional neural networks are typical solution to
the machine learning problems where the input data
has a structure of a finite-dimensional linear space
range. CNNs are biologically inspired (Hubel and
Wiesel, 1965) modification of multilayer perceptron
(MLP) with reduced connections between layers and
extensive weights sharing. One of the most basic
properties is indifference to translation (LeCun and
Bengio, 1995). Unlike MLP, where any permutation
of inputs is equally useful for training, in CNNs the
structure of input data is important and remains pre-
served. Outputs of the hidden layers are called feature
maps (LeCun and Bengio, 1995; Cires¸an et al., 2011),
since they actually describe locations of the certain
features of the image. The CNN input is usually just
a raw digital image, with optional very basic pre-
processing (scaling, normalization, etc.) (Krizhevsky
et al., 2012).
The mentioned properties make CNNs useful for
feature extraction. The usual field where CNNs are
used is image classification – the state-of-art solu-
tions to the ImageNet Large Scale Visual Recogin-
tion Challenge (Deng et al., 2009) are based on CNNs
(Krizhevsky et al., 2012; Zeiler and Fergus, 2013;
Nguyen et al., 2015). However, there are related
works where CNN is used just as general-purpose fea-
ture extractor (Mopuri and Babu, 2015) or as a solu-
tion to the object localization problem (Matsugu et al.,
2003; Dai et al., 2014). Some deep and complex
CNNs trained for ILSVRC were also successfully ap-
plied as a part of larger solution to other image recog-
nition problems (Cheng et al., 2016). The usual ap-
BIOIMAGING 2017 - 4th International Conference on Bioimaging
56

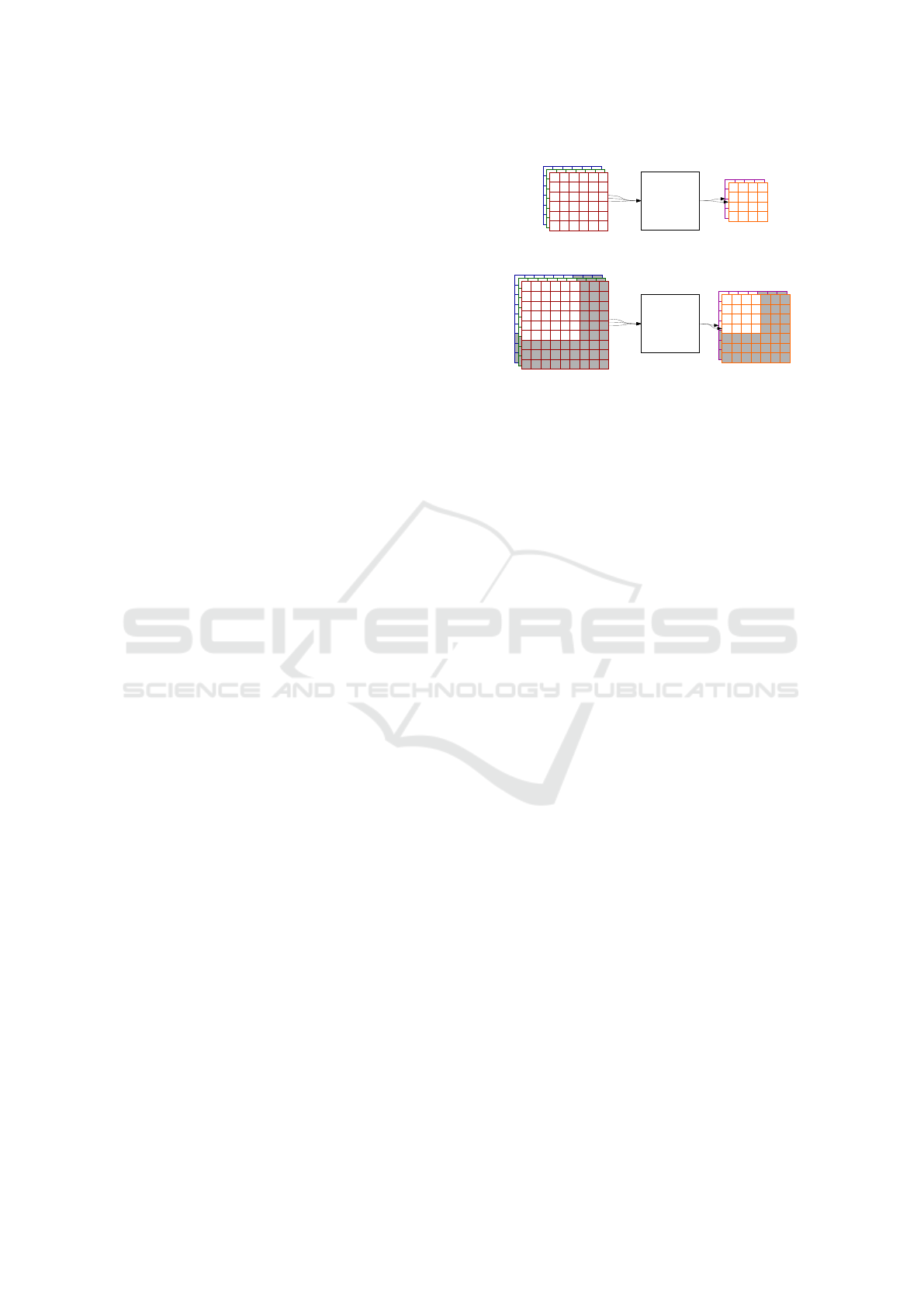
∗
∗
Input
(e.g. 3-channel
image)
Output: one matrix
for each group
of filters
∗
∗
∗
∗
+
+
A
1
, A
2
, A
3
F
1,1
, F
1,2
, F
1,3
F
2,1
, F
2,2
, F
2,3
M
1
, M
2
Figure 1: Convolutional layer internal structure. The exam-
ple setup processes A
1
, A
2
, A
3
input with 2 groups of F
i, j
fil-
ters (3 filters in each group). Convolution results produced
by each filter group are summed up. Each sum is a separate
output matrix, in this case: M
1
, M
2
.
proach expects the CNN to perform some dimension-
ality reduction of the input data, so the size of feature
maps in the consequent hidden layers is decreasing.
The reduced representation calculated with CNN is
usually used with some general-purpose classifier –
MLP is preferred because of easy gradient learning of
the CNN+MLP classifier as a whole (Cires¸an et al.,
2011).
Considering the structure of feature maps, it is
possible to perform a localization task, where the ex-
pected output is a feature map itself. It requires,
however, pure CNN architecture, without a classifier,
since MLP breaks the topological image structure of
the hidden outputs. In order to get a map which could
be easily translated to object location on the input im-
age, we will ensure that the output size is the same as
input size. Instead of decreasing the size of the feature
maps, our approach involves simply keeping them
constant. Detailed application and consequences of
this approach are described in Section 3.2. By nor-
malization of the final CNN output (e.g. with unipo-
lar sigmoid function) we produce a fuzzy map, where
each point is activated according to the likeliness of
belonging to the object. Further processing such as
noise removal and thresholding can be used to get the
binary mask, which is known to be useful in some ap-
plications (Dai et al., 2014). Similar remark applies
to the experiments performed in this work, as it is de-
scribed in Section 4.3. Our approach to thresholding
is presented in Section 3.3.
3.1 Formal Description
Let us denote the input data as a tuple of matrices
A
1
. . . A
p
of a fixed n
a
× m
a
size (for the first layer it
could be multi-channel digital image, or even a single
matrix for p = 1). The key parameters of a convo-
lutional layer (which is the basic unit of CNN) are
q filter groups – each of them being a tuple of p ma-
trices of n
f
× m
f
size (F
i, j
for i = 1 . . . p, j = 1 . . . q).
The output is a tuple of feature maps M
1
. . . M
q
where
for each i = 1 . . . q
M
i
= Z
i
+
p
∑
j=1
!
A
j
∗ F
i, j
.
In the formula above Z
i
is a bias matrix of the same
size as M
i
. Matrix convolution A
j
∗ F
i, j
is a matrix
of elements (A
j
∗ F
i, j
)
r,c
for r = 1 . . . (n
a
)−(n
f
)+1,
c = 1 . . . (m
a
)−(m
f
)+1 such that
(A
j
∗ F
i, j
)
r,c
=
n
f
−1
∑
d
n
=0
!
m
f
−1
∑
d
m
=0
!
(F
i, j
)
(n
f
−d
n
),(m
f
−d
m
)
·
· (A
j
)
(r+d
n
),(c+d
m
)
.
The resulting M
i
matrices size is n
a
−n
f
+1×m
a
−
m
f
+1. Simplified diagram of convolutional diagram
is presented in Fig. 1.
Such a result could be easily processed further
with another convolutional layer. However, since the
matrix convolution with a fixed F
i, j
is linear (and so
is the whole layer), it is advised to use some non-
linearity between the consequent convolutional lay-
ers. The obvious solution is to apply a non-linear ac-
tivation function element-wise. While sigmoid-like
function is known to work, the modern approach is
to use ReLU (rectified linear unit) (Krizhevsky et al.,
2012) or PReLU (parametrized extension of ReLU)
(He et al., 2015). The usual solutions for the clas-
sifier and feature extractor architectures additionally
use maximum- or average-pooling after some of the
convolutional layers. That approach reduces the ma-
trix dimensions by a certain factor (LeCun and Ben-
gio, 1995).
Each element of convolutional layer output is a re-
sult of processing some n
f
× m
f
rectangle picked
from each A
j
. For the first feature map, n
f
× m
f
is
a size of visual field (Hubel and Wiesel, 1965). For
further layers, the size of visual fields could be eas-
ily calculated by tracking down the range of CNN in-
put pixels affecting each output element. Should the
network consist of convolutional layers and element-
wise operations only, the visual field size would
be n
z
× m
z
where n
z
= (n
f
1
+ . . . + n
f
t
) − t + 1 and
m
z
= (m
f
1
+ . . . + m
f
t
) −t + 1. In these formulas t de-
notes a number of convolutional layers and n
f
w
× m
f
w
is w-th layer filter size for w = 1 . . . t.
In our task, where it is desired to keep the orig-
inal size while processing with CNNs, pooling lay-
ers would be counterproductive. For any filters of
size other than 1 × 1 (which would perform just a
Localization of Demyelinating Plaques in MRI using Convolutional Neural Networks
57
point-wise combination), A
j
matrices have different
size than M
i
. To address that problem without any
change to the formulas, we can add padding to the A
j
which would increase the input size to (n
a
+n
f
−1) ×
(m
a
+m
f
−1). Despite the size reduction of the orig-
inal layer, zero-padding can easily prevent any infor-
mation loss. Actually using padding of the proposed
size makes it possible to construct the identity opera-
tor, such as F
i, j
of odd dimensions with 1 in a central
element and 0 everywhere else. Another remark about
flexibility of the proposed solution is that the padding
size (adding (n
f
−1) rows and (m
f
−1) columns) is
independent from the input size – it is related only to
the filter size.
3.2 Detector Training
Our proposed CNN architecture is a superposition of:
zero-padding (of a size which will keep the feature
map size constant) (LeCun et al., 1998), convolutional
layers and element-wise activation functions. We can
train such a network to in order to associate A
1
. . . A
p
with the resulting maps that represent the location of
objects. The location is described in the form of a
binary mask, which contains information about both
position and shape of the detected phenomena. Op-
timally the training images should include not only
the whole object, but some neighboring pixels of the
context as well.
If the object location on the image changes (but
context remains sufficient), the CNN properties auto-
matically guarantee that we will get translated output.
It already makes application of CNN easier, than it
would be for a naive solution which would require
manual application of techniques such as sliding win-
dow. On related note, data augmentation through
small input translations is not necessary with CNNs.
The advantage of CNNs for the described sort of
tasks goes even further than that. Consider image
B
1
. . . B
p
, similar to A
1
. . . A
p
in all terms but size (it
still needs to be the same for each B
j
). It would be
especially practical if B
1
. . . B
p
is just a big image in-
cluding some objects to be detected. In some classi-
cal cases, external solutions such as sliding window
would be considered. Consider using B
1
. . . B
p
as an
input of our CNN. It could be remarked that:
• padding and convolution layer keep the image size
unchanged, since no parameters depend on input
size;
• convolution is still possible to calculate as long
as feature maps are larger than filters (which is
automatically satisfied if B
j
are larger than A
j
);
• element-wise functions are independent of the
map sizes as well.
Input
Output
A
1
, A
2
, A
3
M
1
, M
2
CNN
CNN
B
1
, B
2
, B
3
M’
1
, M’
2
Larger input
Extended output
b)
a)
Figure 2: Consider CNN like in Fig. 1. For each ad-
ditional row/column of input matrices, you get one more
row/column of the output. Case “a)” shows the original
setup. Consider variant “b)”, where the extended input is
used. If you choose a rectangle of the same size as A
j
ma-
trices, some outputs produce results similar to the “a)” setup
(e.g. it would work that way for the pixels of B
j
and M
0
i
dis-
played in white).
The output map would still show the proper mask
of a detected object (Dai et al., 2014), since it was
invariant to translation anyways. Without any addi-
tional utilities – after training on the small samples
(which is remarkably faster than processing a big im-
age with a small object) we get an object detector with
support of any greater input size, as it is shown in
Fig. 2. Detecting multiple objects works out of the
box as well. If there is some space between the ob-
jects to detect, so the visual fields do not intersect, the
process becomes equivalent to the detection of a sin-
gle object.
In order to avoid noise related to the “unknown”
input ranges appearing in the bigger image, our train-
ing set includes negative samples as well, as it is de-
scribed in Section 4.1. Using some context around the
object in the input images already prevents CNN from
picking any points of the included background, but
it leaves the network unprepared for any phenomena
that occur only in greater distance from the detected
objects.
Application of the mentioned methods for de-
myelinating plaques localization is explained further
in Sections 4.1-4.3.
3.3 Evaluation
As mentioned above, the size of the feature maps
is kept constant from layer to layer in the proposed
neural network. We also do not use MLP layers at
the output and the goal of the training is regression
rather than classification. Putting the raw MR scan
BIOIMAGING 2017 - 4th International Conference on Bioimaging
58

on the CNN input we expect that the output consists
of the same-sized image, clearly marking the MS le-
sions as white regions, surrounded by black, neutral
background. In practice, however, the output image
will not be truly black-and-white, and the intensity
of a given output pixel may be interpreted rather in
terms of the probability that it is a part of a lesion.
Therefore, we have to apply thresholding to make the
final decision and to obtain a black-and-white result
that may be directly compared to the expert-generated
ground-truth mask.
The value of the threshold is the fundamental pa-
rameter enabling to control the two elementary mea-
sures of the quality of the results: precision and recall.
Both these measures are based on the count of the
“true-positive” pixels in the CNN output image, i.e.
the pixels with values exceeding the threshold (“pos-
itive”), which at the same time represent the true MS
lesions, as indicated by the ground-truth masks. Re-
call is defined as the proportion of the “true-positive”
pixels to all of the pixels that should be detected (ac-
cording to the mask) and precision is the proportion
of the “true-positive” pixels to all actually detected
pixels. Obviously, low threshold maximizes the re-
call and high threshold maximizes the precision. Ex-
tremely low threshold would render all the pixels pos-
itive, yielding 100% recall and close-to-zero preci-
sion, while extremely high threshold would do the
opposite. Therefore, a standard approach to obtain
a representative results, applied also in our approach,
is to compute the harmonic mean of precision and re-
call, known as F-measure.
The value of F-measure is used in the evaluation
of the obtained results to find the appropriate thresh-
old. We apply a search through all possible thresh-
old values, recording the resulting F-measure values
for the training images. The threshold maximizing
the F-measure is used to compute the final results on
a separate set of testing images, as described in the
following section.
4 EXPERIMENTS
4.1 Dataset Preparation
From the initial set of 100 patients, 4 were removed
from the study due to MR image format discrepan-
cies. The remaining 96 were split at random into the
training set (77 patients) and the testing set (19 pa-
tients). Each patient was represented by a set of MR
scans of the size 448×512 pixels, out of which only
the scans containing plaques of demyelination were
Figure 3: Example of a scan used in the training set (left)
and the accompanying mask (right).
Figure 4: Example of tiles cut from the scan in Fig. 3 (top)
and the accompanying masks (bottom). Note, that tiles b),
e), h) do not contain lesions and that tiles a) and c) represent
the same (topmost) lesion.
considered. As a result, the testing set contained 242
scans and the training set was based on 982 scans.
In the latter case however, the 982 scans were not
directly used but they were cut into tiles of 50×50
pixels and only some of them were selected for in-
clusion into the final training set. Basically, the se-
lected tiles were all those containing demyelinations.
However, preliminary tests revealed that in this way
some parts of the scans, such as the skull bones, areas
around eye globes and sinuses, were never included
in the training set. As an effect, they were usually
mistakenly marked as demyelinations on the testing
set, as they were typically brighter than the surround-
ing regions, similarly to the MS lesions. Therefore, to
let the neural network learn and recognize these areas
and to decrease the risk of false alarms, some of the
tiles without lesions were also included in the training
set (Fig. 3 and Fig. 4). These tiles were selected at
random, but with some additional constraints giving
preference to bright areas and high contrast. Out of
the total number of 7856 tiles constituting the training
set, approximately one-third were these “no-lesion”
tiles. The last operation performed on the tiles with
lesions was to detect when a lesion occurred at the
edge of the tile so that only a part of it was included.
In such cases the tile was shifted appropriately to in-
crease the chance of encompassing the whole lesion.
Localization of Demyelinating Plaques in MRI using Convolutional Neural Networks
59

4.2 CNN Architecture
The structure of the network, i.e. the number of lay-
ers, the number of neurons, the size of the receptive
fields and the non-linearity types were the subject of
intensive experiments in our study. The final archi-
tecture, offering the possibility of successful training
and moderate generalization error is composed of 6
convolutional layers:
• 20 neurons (5×5, padding: 2)
• 20 neurons (7×7, padding: 3)
• 40 neurons (9×9, padding: 4)
• 60 neurons (7×7, padding: 3)
• 20 neurons (5×5, padding: 2)
• 1 neuron (5×5, padding: 2)
We applied parametric rectified linear units (PReLU)
between the layers, and after the last layer the unipo-
lar sigmoid was used.
4.3 Testing Procedure and Results
The experiments were done with Caffe deep learning
framework on a cluster node with Tesla K20M GPU
accelerator. The training set of 7856 50×50 tiles was
fed to the network in mini-batches of 100 tiles each.
Mean square error (Euclidean loss) between the net-
work outputs and the ground-truth masks was used as
the indicator of the training progress.
After several experiments with various parame-
ters of the learning process, we set the initial learn-
ing rate and momentum to 0.00001 and 0.9, respec-
tively. These values guaranteed slow but stable con-
vergence, as demonstrated in Fig. 5 (top plot). The
decrease of the error is clearly visible, which indicates
that the network learns to detect lesions on the train-
ing tiles. It appeared, however, that the enhancement
of the resulting F-measure value was observed only
during the initial phase of the training, as evidenced in
the bottom plot of Fig. 5. We have therefore a typical
problem of generalization error, increasing when the
network is getting overtrained. It should be stressed,
however, that the presented plots correspond to ca. 80
hours of learning, during which the whole training set
was used over 5000 times. It is also worth to note, that
the period of the visible cycles encompasses exactly
625 repetitions of the whole training set
1
.
In the following part, we will demonstrate the
practical effectiveness of the network trained for the
optimal time (ca 15 hours), using the full MR scans
1
This number results from the relation between the size
of the mini-batch and the number of tiles in the training set.
Figure 5: Top: learning curve (Euclidean loss); bottom:
F-measure on the testing set. The unit on the horizontal
axis corresponds to 100 mini-batches (each containing 100
tiles).
448×512 from the testing set. It is worth noting here,
that our CNN composed of convolutional layers only
(no MLP layers) behaves more like an image filter,
accepting any size of the input image without the
need of architecture changes or re-adaptation of the
weights, as pointed out in Section 3.2. This is a sig-
nificant advantage of our approach, enabling to use
full scans to test the network trained on small tiles.
In order to practically verify the effectiveness of
the network on the testing set, we thresholded the net-
work output to obtain the binary image for direct com-
parison with the mask. The value of the threshold was
set so that it maximized the F-measure on the training
set, as described in Section 3.3. It appeared, however,
that the characteristics of the training set composed
of small tiles, was so different from the testing set
containing full scans, that the obtained threshold val-
ues were unsuitable for the use in the testing phase.
Therefore we decided to use the original training im-
ages to compute the threshold. In short, the training
images were used in two forms: cut into tiles (7856
tiles) for network training and uncut (982 scans) for
BIOIMAGING 2017 - 4th International Conference on Bioimaging
60
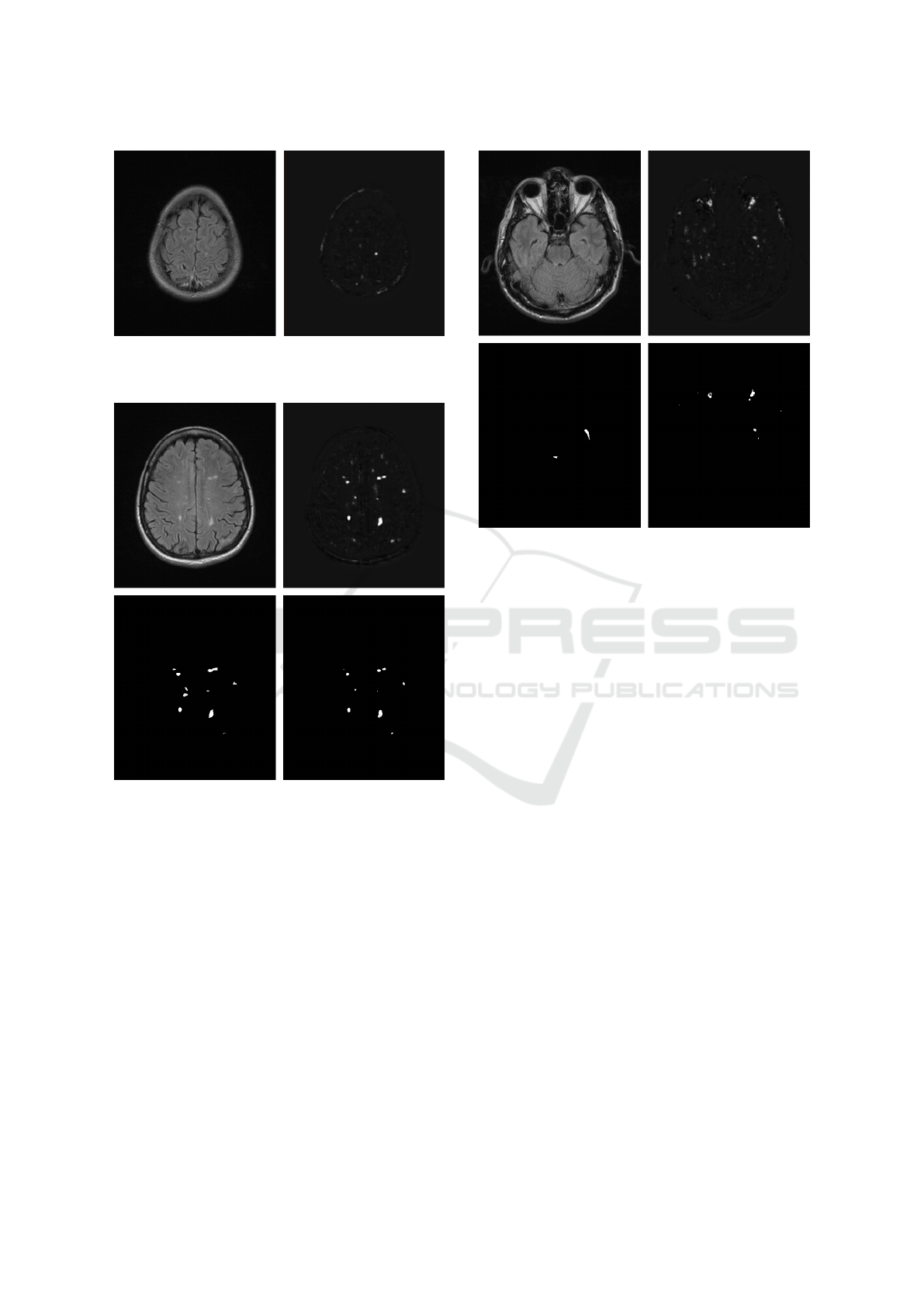
Figure 6: Example of input (left) and output (right) of the
network – a single lesion in the right hemisphere was prop-
erly indicated.
Figure 7: Left column: input image (top) and ground truth
mask (bottom); right column: CNN output image before
thresholding (top) and after thresholding (bottom).
threshold determination.
An example of the results obtained on the testing
set after training is presented in Fig. 6. The image on
the right presents raw network output before thresh-
olding, which in this case, enabled to perfectly de-
tect the demyelination region in the right hemisphere.
It should be noted that this lesion is not very salient
in the input image, which contains many brighter re-
gions, such as the bottom of the hemispheres and the
skull bones. The network actually performs as an
image filter, amplifying the signal in the regions re-
sembling those learned during training, irrespective
of their absolute brightness.
Fig. 7 presents a more complicated case with
many lesions which has also been detected (all except
Figure 8: Left column: input image (top) and ground truth
mask (bottom); right column: CNN output image before
thresholding (top) and after thresholding (bottom).
one in the left hemisphere). In Fig. 8, however, sev-
eral significant problems are revealed, including false
alarms for the tissue surrounding the optic nerves and
the temporal bones. Out of the two genuine lesions
only one is found and unnecessarily split into two dis-
joint regions.
5 ANALYSIS
The examples presented in the previous section give
an idea of what can be expected from our CNN af-
ter training. These results are promising in that they
demonstrate the network’s ability to detect typical de-
myelination lesions. What is important, this ability
seems to be based not only on their intensity but also
on their shape and characteristics of the surrounding
tissue. For comparison, let us consider a simple ap-
proach based on direct thresholding of the raw input
image. An example for the same image as in Fig. 6 is
presented in Fig. 9. As we may observe, the thresh-
old of 50% is too low to correctly detect the lesion,
whereas the number of false positive areas is signif-
icant and it rises dramatically even with quite mod-
erate decrease of the threshold. Clearly, irrespective
on the threshold, the result is virtually useless. On
the other hand, the CNN output presented in Fig. 6
(the image on the right) may be thresholded yielding
the correct outcome, i.e. a single region in the appro-
Localization of Demyelinating Plaques in MRI using Convolutional Neural Networks
61

Figure 9: Example of thresholding of the raw input image
from Fig. 6 with threshold values 50%, 45% and 40% (from
left to right, respectively).
priate location, for a wide range of threshold values
(from 48% to 92% in this particular case).
Considering the overall results it should be noted,
however, that the obtained F-measure values reach
only 55% in the best cases (Fig. 5, bottom) which can-
not be generally deemed a successful outcome. There
are a few sources of this and we will now try to inves-
tigate them in more detail and to formulate possible
solutions.
The first problem, already addressed in Sec-
tion 4.1 is related to the structures adjacent to the
tissues of the central nervous system, such as bones,
meninges and sinuses. They are basically out of the
scope of our study, but they are nevertheless present
in the MR scans so we attempted to purposely train
the CNN to ignore them, as described above. This
attempt was mostly successful, as demonstrated in
Fig. 6 and 7, but it tends to fail in a case when these
structures contain bright regions of considerable size
(Fig. 4). This problem is particularly pronounced in
the topmost MR scans where the bones of the calvaria
are not perpendicular to the projection plane, so they
appear much broader.
There are several possible solutions here, includ-
ing the increase of the number of tiles representing
such structures in the training set, to make the net-
work recognize them better. Another important rem-
edy would also be to increase the size of the tiles.
The 50×50 tiles may be simply too small to incor-
porate enough surrounding tissues in case of bright
regions of significant size. However, both these coun-
termeasures lead to increase of the area without le-
sions in the ground-truth masks. This in turn may
make the network learn to generate purely black out-
put images, because the associated local minimum of
the error function would not differ much from the de-
sired learning goal. The ultimate solution is therefore
to use some automatic or semi-automatic tools to re-
move the irrelevant parts of the input MR scans prior
to CNN training and testing. This would be less uni-
versal, but it would let the CNN concentrate on the
regions of interest (cerebral tissue) only.
The second issue negatively influencing the ob-
tained results is the quality and the quantity of the ma-
terial available for training. Most of the demyelinat-
ing plaques are unambiguously visible in the scans,
but still there are also many small or very faint lesions
which may pose problems in unequivocal identifica-
tion as MS plaques. The problems with generaliza-
tion, indicated in Fig. 5, may suggest that we should
use a significantly bigger set of training images or
more consistently annotated, perhaps by several in-
dependent specialists.
Yet another problem is associated with the preci-
sion of defining the boundaries of the lesions by hu-
man annotators. Quite a significant impact on the
quality of the obtained results stems from the fact
that even if all the lesions were properly detected in
the output image, they typically differ in size and
shape from the ground-truth masks. Due to this fact,
a different approach to the assessment of the out-
comes might be applied: instead of simply count-
ing the matching/non-matching pixels we might only
consider whether a lesion has been detected or not.
This would need some more effort for finding the con-
nected components in the masks and in the thresh-
olded output images, handling the splitting/joining of
adjacent lesions, etc. but evaluation generated in this
way would more accurately reflect the true usefulness
of the obtained results.
6 CONCLUSIONS AND FUTURE
WORK
Careful analysis of brain MRI is an important, time-
consuming part of MS diagnosis. While the final de-
cision on interpretation belongs to the human expert,
artificial intelligence can provide tools that assist the
analysis process. Manual detection and localization
of demyelinating plaques visible on MRI is expected
to be unambiguous, but there is no concise mathemat-
ical formula to describe a plaque. The objective of our
work is to get the best suggestions from the convolu-
tional neural network.
The collected dataset included MRI scans of 100
patients of different age groups. Multiple slices were
stored as relatively large digital images (448 × 512
pixels). Cutting large images into 50 × 50 training
set tiles allowed us to perform the CNN training from
scratch. Due to the CNN properties described in Sec-
tion 3.2, the resulting network supported larger im-
ages out-of-the-box.
In order to compare the result to the target binary
masks properly, a mechanism of automatic threshold-
ing was designed, as it was described in Section 3.3.
As the evaluation was reduced to the comparison of
BIOIMAGING 2017 - 4th International Conference on Bioimaging
62

output and target binary masks, we could directly cal-
culate precision, recall and F-measure.
The best of the proposed models provided F-
measure of 55% on the test set. This value itself
is way from the perfect score. However, getting the
general location of the plaque and slightly imprecise
shape already reduces the value below 100%. The
gold standard consisted of approximate polygons, so
repeating it precisely is virtually impossible. More
significantly problematic factors were related to the
false positives at the large bright areas, such as overly
activated points near the temporal bones and optic
nerves. Another common source of errors was re-
lated to mistakenly activated small regions (noise un-
related to the demyelinating plaques). On the other
hand, presence of selected points in the general area
of demyelinating plaques is a notable advantage of the
suggested model.
This result leaves much room for improvement.
Larger data set, which would include greater vari-
ety of cases, is expected to improve the results. Us-
ing 50 ×50 tiles could be considered disadvantageous
when compared to larger tiles, based on assumption
that larger visual fields could make it easier to recog-
nize temporal bones and optical nerves. However, the
initial tests on larger tiles resulted in all-zero network
outputs, because great majority of target outputs was
black. This problem would have to be addressed by
some specific approach such as cost function modifi-
cation. Another solution could involve creating a sep-
arate tool to remove the irrelevant parts from the im-
age – which means everything besides the brain itself,
where myelin sheath of neurons is visible.
Using convolutional neural networks for medical
image processing is usually difficult because of lim-
ited sizes of data sets. This common problem occured
to our work as well. However, our analysis is a step
towards more efficient solutions. Our approach to the
dynamic threshold selection and chosen measure of
localization correctness (F-measure of the binary ma-
trix) will be useful for testing the future models.
The solutions mentioned above are mostly slight
improvements to the researched method. Another
possible way of the future work involves using pre-
trained CNNs as a part of the model. This is likely
to involve very complex and general solutions such
as AlexNet (Krizhevsky et al., 2012) or VGG (Si-
monyan and Zisserman, 2014). Despite the original
objective of those networks, which is classification,
crucial parts of the same models could be used for
localization as well. Apparently, classification and lo-
calization with CNNs are vastly similar tasks, and one
training process could result in an integrated solution
to both of them (Sermanet et al., 2013). The presence
of the pooling layers results in lower output mask res-
olution. This problem, however, could be addressed
with deconvolutional neural networks (Zeiler and Fer-
gus, 2013).
ACKNOWLEDGEMENTS
This project has been partly funded with support from
National Science Centre, Republic of Poland, deci-
sion number DEC-2012/05/D/ST6/03091.
Authors would like to express their gratitude to the
Department of Radiology of Barlicki University Hos-
pital in Lodz for making head MRI sequences avail-
able.
REFERENCES
Cheng, G., Zhou, P., and Han, J. (2016). Learning rotation-
invariant convolutional neural networks for object de-
tection in vhr optical remote sensing images. IEEE
Transactions on Geoscience and Remote Sensing,
54(12):7405–7415.
Cires¸an, D. C., Meier, U., Masci, J., Gambardella, L. M.,
and Schmidhuber, J. (2011). Flexible, high perfor-
mance convolutional neural networks for image clas-
sification. In Proceedings of the Twenty-Second Inter-
national Joint Conference on Artificial Intelligence -
Volume Volume Two, IJCAI’11, pages 1237–1242.
Dai, J., He, K., and Sun, J. (2014). Convolutional fea-
ture masking for joint object and stuff segmentation.
CoRR, abs/1412.1283.
de Brebisson, A. and Montana, G. (2015). Deep Neural
Networks for Anatomical Brain Segmentation. ArXiv
e-prints, 1502.02445.
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-
Fei, L. (2009). ImageNet: A Large-Scale Hierarchical
Image Database. In CVPR09.
He, K., Zhang, X., Ren, S., and Sun, J. (2015). Delving deep
into rectifiers: Surpassing human-level performance
on imagenet classification. CoRR, abs/1502.01852.
Hubel, D. H. and Wiesel, T. N. (1965). Receptive fields and
functional architecture in two nonstriate visual areas
(18 and 19) of the cat. Journal of Neurophysiology,
28:229–289.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012).
Imagenet classification with deep convolutional neu-
ral networks. In Pereira, F., Burges, C. J. C., Bottou,
L., and Weinberger, K. Q., editors, Advances in Neu-
ral Information Processing Systems 25, pages 1097–
1105. Curran Associates, Inc.
LeCun, Y. and Bengio, Y. (1995). Convolutional networks
for images, speech, and time-series. In Arbib, M. A.,
editor, The Handbook of Brain Theory and Neural
Networks. MIT Press.
LeCun, Y., Bottou, L., Bengio, Y., and Haffner, P. (1998).
Gradient-based learning applied to document recogni-
tion. In Proceedings of the IEEE, pages 2278–2324.
Localization of Demyelinating Plaques in MRI using Convolutional Neural Networks
63

Matsugu, M., Mori, K., Mitari, Y., and Kaneda, Y.
(2003). Subject independent facial expression recog-
nition with robust face detection using a convolutional
neural network. Neural Networks, 16(5-6):555–559.
Milletari, F., Navab, N., and Ahmadi, S.-A. (2016). V-
Net: Fully Convolutional Neural Networks for Vol-
umetric Medical Image Segmentation. ArXiv e-prints,
1606.04797.
Mopuri, K. R. and Babu, R. V. (2015). Object level
deep feature pooling for compact image representa-
tion. CoRR, abs/1504.06591.
Nguyen, T. V., Lu, C., Sepulveda, J., and Yan, S. (2015).
Adaptive nonparametric image parsing. CoRR,
abs/1505.01560.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-Net:
Convolutional Networks for Biomedical Image Seg-
mentation. ArXiv e-prints, 1505.04597.
Sermanet, P., Eigen, D., Zhang, X., Mathieu, M., Fergus,
R., and LeCun, Y. (2013). Overfeat: Integrated recog-
nition, localization and detection using convolutional
networks. CoRR, abs/1312.6229.
Shelhamer, E., Long, J., and Darrell, T. (2016). Fully
Convolutional Networks for Semantic Segmentation.
ArXiv e-prints, 1605.06211.
Simonyan, K. and Zisserman, A. (2014). Very deep con-
volutional networks for large-scale image recognition.
CoRR, abs/1409.1556.
Zeiler, M. D. and Fergus, R. (2013). Visualizing
and understanding convolutional networks. CoRR,
abs/1311.2901.
BIOIMAGING 2017 - 4th International Conference on Bioimaging
64
