
Towards a Bio-inspired Approach to Match Heterogeneous Documents
Nourelhouda Yahi
1
, Hacene Belhadef
1
, Mathieu Roche
2
and Amer Draa
1
1
NTIC Faculty, MISC Laboratory, University of Constantine 2-Abdelhamid Mehri, Algeria
2
UMR TETIS (Cirad, Cnrs, Irstea, AgroParisTech), France
Keywords:
Text Mining, Feature Selection, Semantic Similarity, Quantum Inspired Genetic Algorithm.
Abstract:
Matching heterogeneous text documents coming from different sources means matching data extracted from
these documents, generally structured in the form of vectors. The accuracy of matching directly depends on
the right choice of the content of these vectors. That’s why we need to select the best features. In this paper,
we present a new approach to select the minimum set of features that represents the semantics of a set of text
documents, using a quantum inspired genetic algorithm. Among different Vs characterizing the big data we
focus on ‘Variety’ criterion, therefore, we used three sets of different sources that are semantically similar to
retrieve their best features which describe the semantics of the corpus. In the matching phase, our approach
shows significant improvement compared with the classic ‘Bag-of-words’ approach.
1 INTRODUCTION
In the recent years, the amount of data has been ex-
ploding. Data are generated by different individuals
and originated from different sources. That is why
the format of the presented information is heteroge-
neous and unstructured. Most often, it is expressed
by means of natural language. In order to evaluate the
similarity between documents, usually, a very known
document representation is used which is the ‘Bag-
of-words’ model. This model creates a matrix (i.e.
corpus) with word counts for each data instance (i.e.
documents). The count can be either absolute (i.e.
number of occurrences), binary (contains or does not
contain), sublinear (logarithm of the term frequency),
and so forth. However, if we directly use the vocab-
ulary contained in the training texts, we end up with
a vector space with a very high dimension. Each text
will be represented by a vector with as many terms
as there are words in the vocabulary. The processing
of vectorial space would require a lot of memory and
computation time and could stop us from using more
complex processing algorithms (R
´
ehel, 2005). Fea-
ture selection is a preprocessing technique commonly
used for high-dimensional data. This involves the se-
lection of important feature subset and removing ir-
relevant, redundant and, noisy features, for an easier
and more accurate data presentation.
In the literature, we find different feature selection
methods; but due to the huge increase in the amount
of data, the use of metaheuristics enables us to ob-
tain reasonably good solutions, without needing to
explore the entire solution space. In real-world appli-
cations, people are more interested in obtaining good
solutions in a reasonable amount of time rather than
being obsessed with optimal solutions. Therefore, we
favour metaheuristic methods that have been proven
to be efficient for dealing with real-world applica-
tions (Yusta, 2009). In our experiments we used a
Quantum-Inspired Genetic Algorithm (QIGA) which
offers advantages of genetic algorithms, including the
processing of several data in parallel using the mini-
mum of information, and the providing of a good bal-
ance between exploration and exploitation of research
space, and takes the quantum-inspired algorithms ad-
vantages, including the quantum representation of in-
dividuals; which allows a further exploration; a good
diversity is offered, it can cover a large part of the
search space. Also, quantum algorithms are charac-
terized by a reduced complexity; very few individuals
are necessary for a good representation of the search
space; and a huge computing power thanks to the su-
perposition of states and the quantum operators, en-
abling the processing of a large amount of informa-
tion in parallel (Draa, 2011).
The rest of the paper is organized as follows. We
first discuss previous works related to feature selec-
tion using bio-inspired algorithms in Section 2. We
then describe the proposed approach in Section 3.
Section 4 discusses the details of our experiments and
276
Yahi, N., Belhadef, H., Roche, M. and Draa, A.
Towards a Bio-inspired Approach to Match Heterogeneous Documents.
DOI: 10.5220/0006294002760283
In Proceedings of the 13th International Conference on Web Information Systems and Technologies (WEBIST 2017), pages 276-283
ISBN: 978-989-758-246-2
Copyright © 2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

the results obtained from applying the proposed ap-
proach on heterogeneous documents. Finally, we con-
clude by summarizing the contribution of this work
and giving some perspectives in section 5.
2 RELATED WORK
Because of their advantages, recently, bio-inspired al-
gorithms have been widely used as a tool for feature
selection in data mining. (Kabir et al., 2012) have
proposed a hybrid ant colony optimization (ACO) al-
gorithm for feature selection (FS), called ACOFS, us-
ing a neural network. A key aspect of this algorithm
is the selection of a subset of salient features of re-
duced size. ACOFS uses a hybrid search technique
that combines the advantages of wrapper and filter
approaches. In order to facilitate the hybrid search,
the authors designed new sets of rules for pheromone
update and a heuristic information measurement. On
the other hand, the ants are guided in correct direc-
tions, while constructing graph (subset) paths using a
bounded scheme in each and every step of the algo-
rithm. The above combinations ultimately not only
provide an effective balance between exploration and
exploitation of ants in the search, but also intensify
the global search capability of ACO for a high quality
solution in feature selection. There are other studies
that applied the ant colony algorithm to the problem
of feature selection such as (Al-Ani, 2005) and (Agh-
dam et al., 2009).
(Zahran and Kanaan, 2009) have introduced a fea-
ture selection algorithm based on Particle Swarm Op-
timization (PSO) to improve the performance of Ara-
bic text categorization. They used RBF networks (Ra-
dial Basis Function) as a text classifier. On the basis
of the same bio-inspired algorithm, (Xue et al., 2012)
have proposed two multi-objective algorithms for se-
lecting the Pareto front of non-dominated solutions
(feature subsets) for classification. The first algorithm
introduces the idea of non-dominated sorting based
multi-objective genetic algorithm into PSO for feature
selection. In the second algorithm, the multi-objective
PSO uses the ideas of crowding, mutation and domi-
nance to search for the Pareto front solutions.
(Siedlecki and Sklansky, 1989) introduced the use
of Genetic Algorithm (GA) for feature selection. In
a GA approach, a given feature subset is represented
as a binary string ‘Chromosome’ of length n, with a
zero or one in a position i denoting the absence or
presence of feature i in the set, respectively. Note that
n is the total number of available features. A popu-
lation of chromosomes is maintained. Each chromo-
some is evaluated to determine its ”fitness”, which de-
termines how likely the chromosome is to survive and
breed into the next generation. New chromosomes
are created from old chromosomes by the following
processes: (1) crossover, where parts of two differ-
ent parent chromosomes are mixed to create offspring
and (2) mutation, where the bits of a single parent are
randomly disturbed to create a child (Yusta, 2009).
(Jourdan et al., 2001) also presented a genetic algo-
rithm dedicated for a feature selection problem, but in
a particular case encountered in the genetic analysis
of different diseases. The specificities of this problem
is that the authors are not looking for a single feature,
but for several associations of features that may be in-
volved in the studied disease. There are other studies
applying the genetic algorithm on the problem of fea-
ture selection, we cite those of: (Yang and Honavar,
1998), (Oliveira et al., 2003) and (Babatunde et al.,
2014).
3 THE PROPOSED APPROACH
The proposed approach is composed of three mod-
ules: preprocessing, feature selection and matching.
Figure 1 shows the structure of the proposed ap-
proach. First, for the preprocessing of the input het-
erogeneous text documents, we implement the most
important prerocessing steps which include opera-
tions such as cleaning data and stemming. Second,
we apply a quantum-inspired genetic algorithm in or-
der to select the minimum set of features that are ide-
ally necessary and sufficient to describe the semantics
of a set of heterogeneous text documents; in order to
reduce the cost and increase the matching accuracy of
these documents. Finally, the cosine similarity is used
to measure the difference between the input text doc-
ument and its corresponding in the matching phase.
The following subsections describe the details of each
step of the proposed approach.
3.1 Preprocessing Phase
Text preprocessing is a task that plays a very impor-
tant role in text mining techniques and applications,
it becomes even more important when handling big
data generated from multiple sources. In this step,
we used R language which is widely used among data
miners, it offers multiple packages for performing text
mining that facilitate preprocessing tasks including:
the elimination of punctuation, digits and stopwords,
stemming, TF-IDF weighting, etc.
As our bio-inspired approach is intended to deal
with heterogeneous text documents, the collected
dataset used in our work consists of scientific articles,
Towards a Bio-inspired Approach to Match Heterogeneous Documents
277
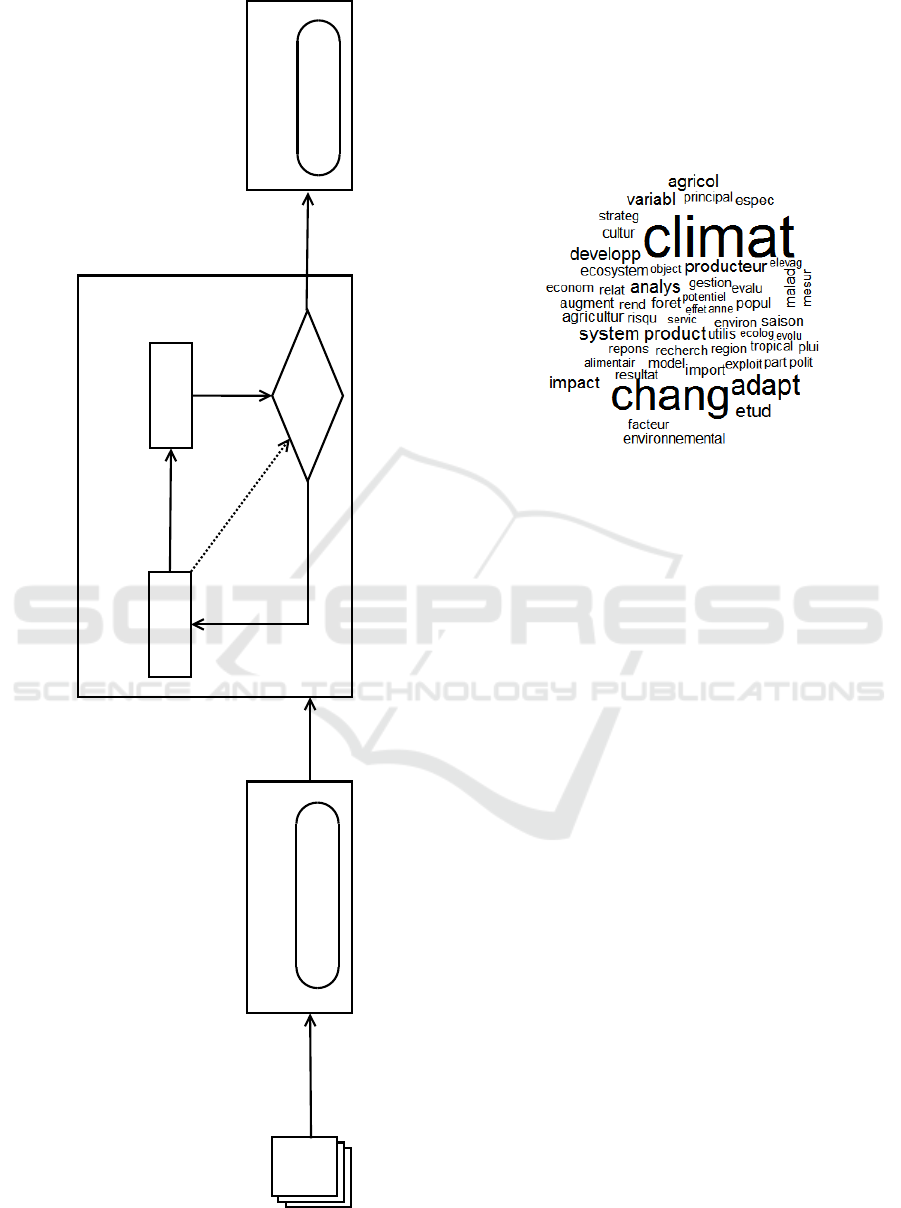
Dataset
Heterogeneous
Documents
Preprocessing
Cleaning and stemming
Feature
vectors
Criterion
Generation
Evaluation
Features
subset
Subset
quality
No
Feature selection with QIGA
Yes Selected
features
Matching
Cosine similarity
Figure 1: The proposed bio-inspired approach to match het-
erogeneous documents.
blog posts, and tweets. In order to clean the corpus
of scientific articles and blog posts, the tasks carried
out are: removing links, punctuation and digits, re-
moving stopwords using a standard list that we have
enriched, eliminating unnecessary spaces, stemming,
and deemphasis. Concerning tweets, we have added
for them removing hashtags and citations.
Figure 2: Word cloud of a preprocessed corpus.
3.2 Feature Selection Phase
The excessive dimensionality of feature vectors in-
creases the training time and decreases matching ac-
curacy. In order to select the best features, we
chose to use the Quantum-Inspired Genetic Algo-
rithm (QIGA); to take advantage from the genetic
algorithm which is suitable for discrete optimisation
problems and from quantum computing, which is able
to minimize the complexity of the algorithm.
3.2.1 Quantum-inspired Genetic Algorithm
A quantum-inspired genetic algorithm is a genetic al-
gorithm enriched by the concepts and principles of
quantum computing, such as the qubit, the superposi-
tion of states and the quantum operators. In quantum
computing, the smallest unit of information storage is
the qubit. A qubit can be in the state 1, the state 0 or
in a superposition of both. The state of a qubit can be
represented as indicated by Formula (1).
|Ψi = α|0i + β|1i (1)
Where |0i and |1i represent the conventional values
of bits 0 and 1, respectively. α and β are complex
numbers satisfying:
|α|
2
+ |β|
2
= 1 (2)
|α|
2
represents the probability that the qubit is found
in the state 0, while |β|
2
represents the probability that
the qubit is found in the state 1. A quantum register of
WEBIST 2017 - 13th International Conference on Web Information Systems and Technologies
278
m bits can represent 2
m
values simultaneously. How-
ever, in the act of observing a quantum state, there is
no more superposition and one of the values is then
available for use (Laboudi and Chikhi, 2009). As the
basic element here is the qubit, a chromosome is sim-
ply a string of m qubits forming a quantum register.
To extract a classical chromosome from a quan-
tum chromosome, we apply the quantum measure-
ment, measuring the state of a qubit forces it to point
to either ‘1’ or to ‘0’. The result depends on the am-
plitudes of the qubit, such as a qubit whose value
|α|
2
= 0.8 will have an 80% chance of being a ‘1’
and 20% to be in the state ‘0’.
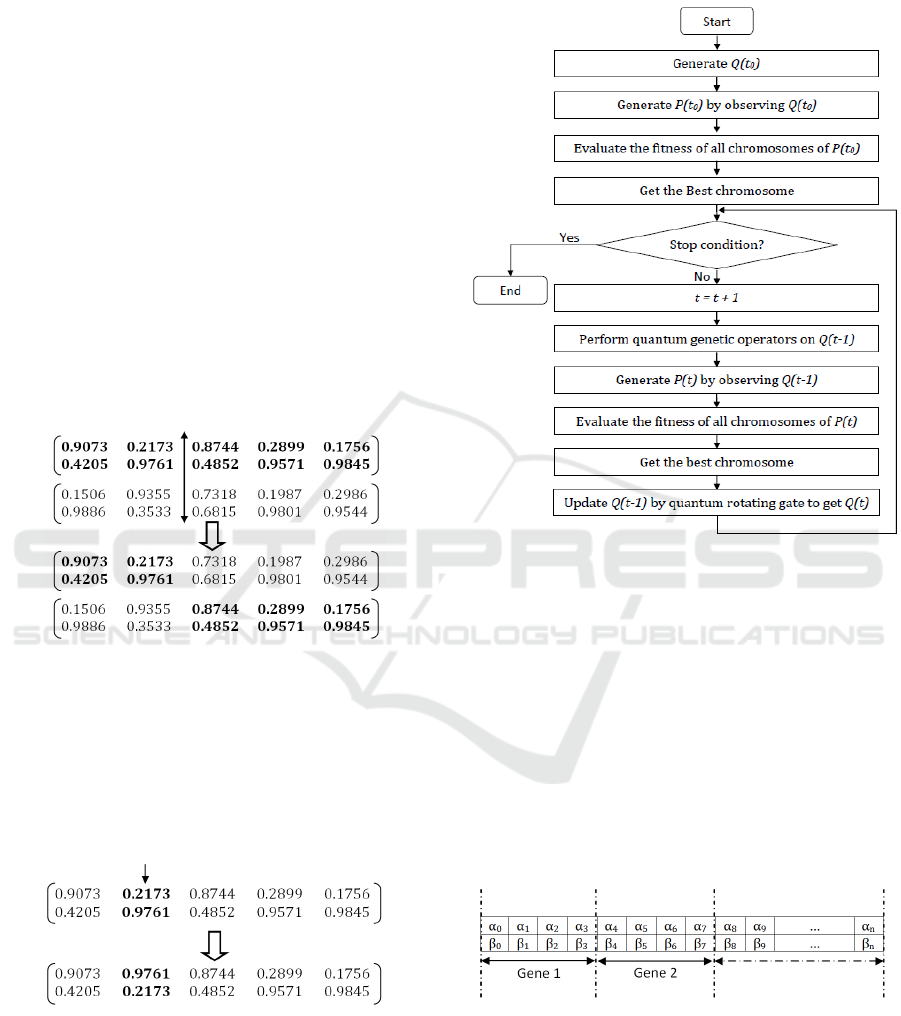
Quantum crossover has the same principle as a
conventional crossover. But it operates on quan-
tum chromosomes. So, it is a probability matrice
crossover that generates as a result new probabil-
ity matrices. The quantum crossover between two
individuals (parents) at a given point can generate
two new individuals (offspring) whose genes become
from both parents. As shown in the Figure 3.
Figure 3: Quantum crossover.
Classical mutation operates as a small perturba-
tion that reverses the mutated bit. In a quantum mu-
tation, there is also a perturbation, but it operates on
the probabilities of a qubit of the concerned chromo-
some, as follows. Consider a qubit |Ai = α|0i + β|1i.
The qubit quantum mutation of A generates the qubit
|Bi = β|0i + α|1i.
Figure 4: Quantum mutation.
At each iteration, the current best solution serves
as a guide to find new solutions that may be better.
This is implemented via a quantum gate, which ro-
tates the qubit in question in order to increase the
probability of having the binary value of the corre-
sponding bit in the current best solution (Draa, 2011).
The general principle of the quantum-inspired genetic
algorithm is illustrated in Figure 5.
Figure 5: Flowchart of the quantum inspired genetic algo-
rithm.
3.2.2 Representation of Solutions
In our bio-inspired approach a quantum representa-
tion of solutions is adopted. A given feature subset is
represented as a quantum register ‘Quantum chromo-
some’ of length n, where each qubit is composed of
two values, namely α and β in position i denoting the
absence or presence of feature i in the set. Note that
n is the total number of available features. A popula-
tion is simply a set of quantum chromosomes. Figure
6 shows the structure of a quantum chromosome.
Figure 6: The quantum chromosome structure.
3.2.3 Objective Function
In general, the objective function consists of two
terms which are in competition with each other: the
number of features (to be minimized) and the quality
Towards a Bio-inspired Approach to Match Heterogeneous Documents
279

(to be maximized). The decision is a compromise be-
tween these two objectives. In our approach the qual-
ity is evaluated with the cosine measure between the
vector of features to reduce and the learning vectors.
Given two n dimensional vectors ~v and ~w , the cosine
similarity between them is calculated as follows:
Cosine(~v,~w) =
~v ·~w
|~v||~w|
=
∑
n
i=1
v
i
× w
i
q
∑
n
i=1
v
2
i
q
∑
n
i=1
w
2
i
(3)
The resulting similarity ranges from 0 meaning totally
different, to 1 meaning exactly the same. In order to
calculate the cosine similarity we must pass by the
weighting step. In which we used two methods, the
first method consists of concatenating the two feature
vectors (i.e. the vector to be optimized, and the vec-
tor used for learning). Then, two weight vectors are
constructed in this way: for each feature, if it does not
exist in the vector to optimize (for P1, but for P2 the
existence of the term is verified in the learning vector)
the given weight is ‘0’, otherwise, if it exists in the
vector to optimize only the given weight is ‘1’, oth-
erwise, if it exists in the two vectors the given weight
is ‘2’. The second weighting method is based on the
TF-IDF measure (Salton and McGill, 1986) which is
a weighting function that depends on the term fre-
quency (TF) in a given document calculated with its
relative collection frequency (IDF).
Since the calculation of TF-IDF for each term in
relation to each document takes a lot of time, in our
approach the TF-IDF value of each term is calculated
in relation to each corpus. Then, the fitness is calcu-
lated using Formula (4).
Fitness = α ∗ Size + (1 − α) ∗ Quality (4)
Data: Quantum chromosomes of features to reduce
+ Chromosomes of learning features
Result: fitness of each chromosome
for each quantum chromosome do
Apply quantum measurement
Calculate the number of non-selected features:
zeros
Concatenate the feature vectors (selected +
learning)
Generate two weight vectors: P1 and P2
Calculate the cosine similarity between P1 and
P2: cos
Calculate Fitness = α * zeros + (1-α) * cos
end
Algorithm 1: The objective function.
3.3 Matching Phase
In order to match heterogeneous text documents, we
calculate the cosine similarity using two methods, the
first one consists in calculating the cosine between
vectors representing the terms occurrences, and the
second is a cosine calculation between TF-IDF vec-
tors (Term Frequency-Inverse Document Frequency)
of all test pairs at the basis of the features selected
from the previous phase.
4 EXPERIMENTAL RESULTS
In this section, the proposed approach is validated
through applying it on three types of text documents:
scientific articles, blog posts, and tweets. Then MRR
(Mean Reciprocal Rank) measurement is adopted in
order to compare the proposed approach to the classic
‘Bag-of-words’ approach.
4.1 Corpora
In this work, we focus on ‘Variety’ criterion of big
data. Consequently, we have collected heterogeneous
documents in French from various sources.
• Scientific articles dealing with climate change
subject from an abstract collection of arti-
cles, books, book chapters, thesis, etc., gotten
from: Agritrop open archive CIRAD
1
(Agri-
cultural research and international cooperation,
French organization working for the sustainable
development of tropical and Mediterranean re-
gions’publications). It offers free access to writ-
ten scientific production of CIRAD in compliance
with copyright. In 2016, Agritrop includes more
than 92,000 references and 25,000 full-text docu-
ments in agricultural research and rural develop-
ment in Southern countries.
• Non-climatic scientific articles from a collection
of abstracts of TETIS
2
Laboratory (for Territories,
Environment, Remote Sensing and Spatial Infor-
mation). It aims to develop methods of spatial in-
formation control to promote the knowledge and
management of environment and territories. The
research domains of these paper are agro-forestry,
urbanization, natural resource management, land-
use planning, etc.
• Blog posts addressing the climate issue that are
manually collected from two blogs
3
that address
the theme of climate change.
1
http://www.cirad.fr/
2
https://tetis.teledetection.fr/index.php/fr/tetis-summary
3
http://www.ecoco2.com/blog/rechauffement-
climatique and http://maplanete.blogs.sudouest.fr/tag/
rechauffement+climatique
WEBIST 2017 - 13th International Conference on Web Information Systems and Technologies
280

• Non-climatic blog posts that are manually col-
lected from a blog
4
of useful information of
medicines called non-conventional or alternative.
• Tweets dealing with climate change topic from
DEFITweets
5
which is a corpus of climate change
(15,000 tweets).
• Non-climatic tweets from Politweets corpus
(Longhi et al., 2014). This corpus gathers the
tweets of 7 personalities from 6 different French
political groups. Extracted from Twitter accounts
of these persons by a method that selects mes-
sages sent in 2013 and 2014, making the total cor-
pus of 34273 messages (tweets).
In our experiments we have randomly selected 100
documents for each category. These data are named:
CA for scientific Articles in Climate change domain,
NCA represents Non-Climatic scientific Articles, CB
for Climatic Blog posts, NCB for Non-Climatic Blog
posts, CT represents Climate change Tweets and NCT
is for Non-Climatic Tweets, where relevant pairs are
pairs of heterogeneous documents covering the same
subject; e.g. a document from CA and a document
from CB.
4.2 Experimental Protocol
After data preprocessing our goal is to select the
minimum features that describe the semantics of the
corpus using the quantum-inspired genetic algorithm
with the following parameters. The number of it-
erations has been fixed at 300, the population size
was chosen to be 5, the probability of mutation and
crossover were chosen to be 0.1 and 0.9, respectively.
The number of documents for each category is 66
and the α value in the objective function took values
from 0.1 to 0.9. The quantum-inspired genetic algo-
rithm proceeds as follows. To extract the best features
of climate Articles CA, the Algorithm inputs are CA
feature vector as the vector to be reduced and (CB
+ CT) feature vector as the vector for learning fea-
tures. First, we start by randomly creating the initial
population. Then, for each iteration we apply the ge-
netic operators, we evaluate the population using the
formula of the objective function, and we extract the
words chosen by the algorithm. After that, we ap-
ply the above steps to extract the best features of cli-
mate blogs (inputs are CB to reduce and (CA + CT)
as learning vector), and the best features of climate
tweets (inputs are CT to reduce and (CA + CB) as
a learning vector). Finally, the base of features that
4
http://blog-medecine-douce.com/
5
https://deft.limsi.fr/2015/index.php
describes the semantics of the corpus is the concate-
nation of the best features selected in CA, CB and CT.
4.3 Mean Reciprocal Rank
The Reciprocal Rank (RR) information retrieval mea-
sure calculates the reciprocal of the rank at which the
first relevant document was retrieved. RR equals 1
if a relevant document was retrieved at rank 1, if not
it equals 0.5 if a relevant document was retrieved at
rank 2 and so on. When averaged across queries, the
measure is called the Mean Reciprocal Rank (MRR)
(Craswell, 2009). This statistical measure evaluates
any process that produces a list of possible answers
to a sample of queries, ordered by the probability of
correctness. In our experiments, MRR is calculated in
two ways, the first way is based on the calculation of
the cosine between term-occurrence vectors of all test
pairs, and the second one is based on the calculation
of the cosine between TF-IDF vectors of all test pairs.
These vectors are calculated on the basis of the fea-
tures selected with the quantum-inspired genetic algo-
rithm. The cosine values are sorted in descending or-
der, then couples relevance is evaluated, knowing that
relevant pairs are pairs of heterogeneous documents
covering the same subject; e.g. a document from CA
and a document from CB, by assigning the value 1 to
the relevant couples and 0 for irrelevant ones.
Table 1: Reciprocal Rank calculation based on cosine simi-
larity.
ID Doc 1 ID Doc 2 Cosine Relevance P@i
166 11 1.000 1 1
58 64 0.944 1 1
16 65 0.914 0 2/3
120 2 0.810 1 3/4
... ... ... ... ...
After that, for each couple i, precision P@i is cal-
culated as follows; If r relevant documents have been
retrieved at rank i, then:
P@i =
r
i
(5)
Finally we calculate the mean reciprocal rank us-
ing Formula 6.
MRR =
1
n
n
∑
i=1
P@i (6)
4.4 Results
We present in this section the results obtained from
applying the proposed bio-inspired approach on three
categories of heterogeneous text documents collected
from different sources which are; scientific articles,
Towards a Bio-inspired Approach to Match Heterogeneous Documents
281
blog posts, and tweets. These results are compared to
those given by the classic ‘Bag-of-words’ approach.
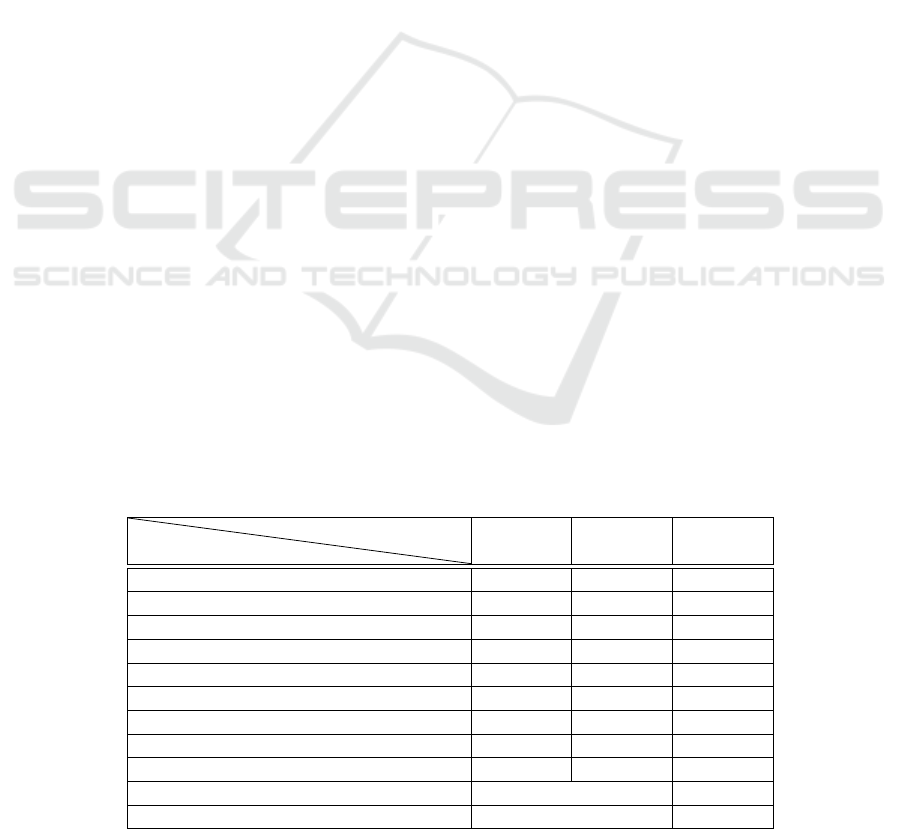
Table 2 shows the results of three variations of the
proposed approach, which are different in two steps;
the weighting step which precedes the cosine calcu-
lation in the objective function, and the cosine calcu-
lation step that precedes the MRR calculation. The
first method uses a proposed weighting method, and
the MRR is calculated using the occurrence vectors
of the terms of all test pairs. The second method uses
TF-IDF as a weighting measure, and the MRR is cal-
culated using the occurrence vectors of the terms of
all test pairs. The last method uses the TF-IDF as a
weighting measure, and the MRR is calculated using
the TF-IDF vectors of all the test pairs. MRR values
are calculated for:
• The feature bases constructed by different α val-
ues of the objective function: Fitness = α ∗Size +
(1 − α) ∗ Quality.
• The features of learning corpus, after the prepro-
cessing phase.
• The features of learning corpus without prepro-
cessing, only punctuation removing.
From the reported results, we can see that the
first method did not give better results compared to
the preprocessed ‘Bag-of-words’ approach, but it out-
performed the not preprocessed ‘Bag-of-words’ ap-
proach for three alpha values (0.2, 0.4, and 0.6). The
second method outperformed the preprocessed ‘Bag-
of-words’ approach for two alpha values (0.1 and 0.6)
and the not preprocessed ‘Bag-of-words’ approach
for all alpha values. In the third method the results
of the three alpha values (0.1, 0.2, and 0.6) were bet-
ter compared to the preprocessed ‘Bag-of-words’ ap-
proach, and the not preprocessed ‘Bag-of-words’ was
worse compared to all other results.
5 CONCLUSION AND FUTURE
WORK
In this paper, we have proposed a bio-inspired ap-
proach for matching heterogeneous text documents.
In the first stage, we preprocess data by cleaning and
stemming to get a feature vector. The second phase
highlights the selection of the minimum set of fea-
tures that represents the semantics of text documents
using the quantum-inspired genetic algorithm. On the
basis of the features selected in the previous phase,
we do the matching. In order to validate the proposed
approach, three document sets coming from differ-
ent sources, that are semantically similar, are used
to retrieve their optimal features. Then MRR mea-
surement is used to evaluate matching accuracy. The
proposed approach outperformed the classical ‘Bag-
of-words’ approach.
Our future work intends to apply other weighting
measurements that can bring further improvements as
Okapi BM25, we would also compare the proposed
approach with other works using other corpora.
ACKNOWLEDGEMENT
This work has been supported by SONGES project
(FEDER and Occitanie).
REFERENCES
Aghdam, M. H., Ghasem-Aghaee, N., and Basiri, M. E.
(2009). Text feature selection using ant colony
optimization. Expert systems with applications,
36(3):6843–6853.
Al-Ani, A. (2005). Ant colony optimization for feature sub-
set selection. In WEC (2), pages 35–38. Citeseer.
Table 2: Comparison of matching accuracy of the proposed approach with different objective functions.
Objective function
ID Method
Method 1 Method 2 Method 3
0.1 * Size + 0.9 * Quality 0.7953 0.9578 0.9914
0.2 * Size + 0.8 * Quality 0.9319 0.9497 0.9865
0.3 * Size + 0.7 * Quality 0.8640 0.9171 0.9247
0.4 * Size + 0.6 * Quality 0.9190 0.9439 0.9706
0.5 * Size + 0.5 * Quality 0.8751 0.9361 0.9461
0.6 * Size + 0.4 * Quality 0.8741 0.9534 0.9963
0.7 * Size + 0.3 * Quality 0.8682 0.9461 0.9759
0.8 * Size + 0.2 * Quality 0.9274 0.9171 0.9247
0.9 * Size + 0.1 * Quality 0.8643 0.9439 0.9706
Bag-of-words (preprocessed) 0.9512 0.9770
Bag-of-words (not preprocessed) 0.8898 0.7920
WEBIST 2017 - 13th International Conference on Web Information Systems and Technologies
282

Babatunde, O., Armstrong, L., Leng, J., and Diepeveen, D.
(2014). A genetic algorithm-based feature selection.
British Journal of Mathematics & Computer Science,
4(21):889–905.
Craswell, N. (2009). Mean reciprocal rank. In Encyclopedia
of Database Systems, pages 1703–1703. Springer.
Draa, A. (2011). Mod
´
eles pour les syst
´
eemes complexes
adaptatifs pour la r
´
esolution de probl
´
emes : Auto-
mates cellulaires apprenants quantiques et
´
evolution
diff
´
erentielle quantique. PhD thesis, Constantine 2
Abdelhamid Mehri University, Algeria.
Jourdan, L., Dhaenens, C., and Talbi, E.-G. (2001). A ge-
netic algorithm for feature selection in data-mining for
genetics. Proceedings of the 4th Metaheuristics Inter-
national ConferencePorto (MIC2001), pages 29–34.
Kabir, M. M., Shahjahan, M., and Murase, K. (2012).
A new hybrid ant colony optimization algorithm for
feature selection. Expert Systems with Applications,
39(3):3747–3763.
Laboudi, Z. and Chikhi, S. (2009). Evolution d’automate
cellulaire par algorithme genetique quantique. In
CIIA.
Longhi, J., Marinica, C., Borzic, B., and Alkhouli, A.
(2014). Polititweets, corpus de tweets provenant
de comptes politiques influents. Banque de cor-
pus CoMeRe. Ortolang. fr: Nancy. http://hdl. handle.
net/11403/comere/cmr-polititweets.
Oliveira, L. S., Sabourin, R., Bortolozzi, F., and Suen,
C. Y. (2003). A methodology for feature selec-
tion using multiobjective genetic algorithms for hand-
written digit string recognition. International Jour-
nal of Pattern Recognition and Artificial Intelligence,
17(06):903–929.
R
´
ehel, S. (2005). Cat
´
egorisation automatique de textes et
cooccurrence de mots provenant de documents non
´
etiquet
´
es. Faculty of Science and Engineering, Uni-
versity LAVAL, QUEBEC.
Salton, G. and McGill, M. J. (1986). Introduction to modern
information retrieval.
Siedlecki, W. and Sklansky, J. (1989). A note on genetic
algorithms for large-scale feature selection. Pattern
recognition letters, 10(5):335–347.
Xue, B., Zhang, M., and Browne, W. N. (2012). Multi-
objective particle swarm optimisation (pso) for feature
selection. In Proceedings of the 14th annual confer-
ence on Genetic and evolutionary computation, pages
81–88. ACM.
Yang, J. and Honavar, V. (1998). Feature subset selection
using a genetic algorithm. In Feature extraction, con-
struction and selection, pages 117–136. Springer.
Yusta, S. C. (2009). Different metaheuristic strategies to
solve the feature selection problem. Pattern Recogni-
tion Letters, 30(5):525–534.
Zahran, B. M. and Kanaan, G. (2009). Text feature se-
lection using particle swarm optimization algorithm.
World Applied Sciences Journal 7 (Special Issue of
Computer & IT), pages 69–74.
Towards a Bio-inspired Approach to Match Heterogeneous Documents
283
