
How to Disassemble a Virus Capsid
A Computational Approach
Claudio Alexandre Piedade, Ant
´
onio E. N. Ferreira and Carlos Cordeiro
Laborat
´
orio de FTICR e Espectrometria de Massa Estrutural, Centro de Qu
´
ımica e Bioqu
´
ımica,
Departamento de Qu
´
ımica e Bioqu
´
ımica, Faculdade de Ci
ˆ
encias, Universidade de Lisboa, 1749-016 Lisboa, Portugal
Keywords:
Virus, Capsid Disassembly, Combinatorial Geometry, Symmetry Groups, Structural Biology.
Abstract:
In contrast with the assembly process of virus particles, which has been the focus of many experimental and
theoretical studies, the disassembly of virus protein capsids, a key event during infection, has generally been
overlooked. Although the nature of the intracellular triggers that promote subunit disassembly may be diverse,
here we postulate that the order of subunit removal is mainly determined by each virus structural geometry
and the strength of subunit interactions. Following this assumption, we modelled the early stages of virus
disassembly of T = 1 icosahedral viruses, predicting the sequence of removal of up to five subunits in a sample
of 51 structures. We used combinatorics and geometry, to find non-geometrically identical capsid fragments
and estimated their energy by three different heuristics based on the number of weak inter-subunit contacts.
We found a main disassembly pathway common to a large group of viruses consisting of the removal of a
triangular trimer. Densoviruses lose a square-shaped tetramer while Human Adenoviruses lose a pentagon-
shaped pentamer. Results were virtually independent of the heuristic measure used. These findings suggest
that particular subunit interactions might be an important target for novel antiviral drugs designed to interfere
with capsid disassembly.
1 INTRODUCTION
Viruses are intracellular parasites that replicate inside
living cells, by using its genetic and protein synthesis
machinery to create new copies (Poranen et al., 2002;
Mateu, 2013). Viral particles are composed by a nu-
cleic acid (DNA or RNA, single or double stranded)
and, in many cases, a capsid, a proteic structure that
protects the genetic information in between infections
(Caspar and Klug, 1962; Poranen et al., 2002; Prasad
and Schmid, 2012). Round-shaped viruses have a
fixed number of proteins or asymmetric units sur-
rounding the genetic material in a icosahedral sym-
metry (Caspar and Klug, 1962; Prasad and Schmid,
2012). This arrangement was first described by Cas-
par and Klug in 1962, who developed a method to
classify the icosahedral symmetry by triangulating the
icosahedron facets. The Triangulation number (or T -
number) represents the number of equilateral trian-
gles that compose a triangular face of the icosahedron
(Prasad and Schmid, 2012).
Many studies have addressed the virus assembly
process, both experimentally and theoretically (re-
viewed in (Perlmutter and Hagan, 2015) and (Mateu,
2013)). Viruses’ capsid subunits are held together by
non-covalent interactions (Zlotnick, 1994; Zlotnick
et al., 1999; Zlotnick, 2003) such as electrostatic salt
bridges, hydrophobic contacts and hydrogen bonds.
The effects of a myriad of these weak interactions is
a globally stable capsid (Zlotnick and Stray, 2003).
Virus assembly is spontaneous in vitro under close-
to-physiological conditions (Mateu, 2013) and the re-
sulting capsids are stable for a long period of time.
Results derived from theoretical models of virus as-
sembly consistently agree on the prediction that three
trimeric protomers, pentamers of trimers and capsids
with just a triangular icosahedron face missing are
stable intermediate forms that precede the formation
of a complete capsid (Zlotnick, 1994; Zlotnick et al.,
1999; Reddy et al., 1998; Reddy and Johnson, 2005;
Rapaport, 2008; Rapaport, 2010). If capsid disassem-
bly was just the reverse process of capsid assembly,
these finding would suggest that the same structures
would be found during disassembly. In high contrast
to the number of studies focusing on the assembly
process, there are very few experimental studies on
capsid disassembly pathways. Since it is expected
that intermediates of this process are transient and
very difficult to detect experimentally, models of the
disassembly pathways are always difficult to confirm
Piedade C., Ferreira A. and Cordeiro C.
How to Disassemble a Virus Capsid - A Computational Approach.
DOI: 10.5220/0006249802170222
In Proceedings of the 10th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2017), pages 217-222
ISBN: 978-989-758-214-1
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
217

experimentally. Moreover, the assembly and disas-
sembly pathways might not be symmetric since some
viruses undergo maturation steps after assembly com-
pletion, such as proteolysis, cross-linking or confor-
mational change (Zlotnick and Stray, 2003). Castel-
lanos et al. (Castellanos et al., 2012), using Atomic
Force Microscopy (AFM), observed the removal of a
triangle of subunits (an icosahedral face) when force
was applied over the Minute Virus of Mice, followed
sometimes by the loss of an adjacent triangle. In
other cases, a removal of a pentamer of triangles (15
proteins) was observed. This follows the predictions
of the assembly models (Reddy et al., 1998; Reddy
and Johnson, 2005; Rapaport, 2008; Rapaport, 2010).
Horton and Lewis (Horton and Lewis, 1992) stated
that the energy minimums appear every multiple of
three subunits. Ortega-Esteban et al. (Ortega-Esteban
et al., 2013) have shown also by AFM that for Hu-
man Adenoviruses the disassembly, both for mature
and immature capsids, start by the loss of a pentagon
of proteins.
The work described here tackles the lack of the-
oretical studies targeting the disassembly process re-
lying on rigorous geometrical and combinatorial con-
siderations, focusing primarily on T = 1 icosahedral
capsid structures. This work in progress has the long-
term goal of providing insights into the disassembly
process, which can be used in the development of an-
tiviral drugs interfering with intermediate states of the
process.
2 METHODS
2.1 Combinatorics and Symmetry
Virus capsid structures with T = 1, although hav-
ing icosahedral symmetry, are not icosahedron-like,
as they contain 60 proteins instead of only 20. The
60-face polyhedron, in which each triangular face of
the icosahedron is divided into three faces, is the Del-
toidal Hexecontahedron. Due to the symmetry of the
Deltoidal Hexecontahedron, the combinations of re-
moving N proteins from a capsid (
60
N
) leads to re-
dundancy since some are symmetrically equivalent,
and the real number is smaller. The exact number of
combinations can be obtained by studying the Burn-
side’s Lemma (Burnside, 1909). To build up the
group of permutations associated with the rotations
of a Deltoidal Hexecontahedron, the faces were num-
bered as shown in Figure 1, followed by the analy-
sis of the transformations done on the numbering of
the faces by the symmetry operators of the symme-
try group I (Vincent, 2001). Applying the different
Figure 1: Graph representation of the Rhombicosidodeca-
hedron. Each vertex represents a subunits of the T = 1 virus
capsid. Edges represent geometrical edges.
rotations on the symmetry axes of a Deltoidal Hex-
econtahedron, results in a permutation group. These
permutations can be seen as permutations of faces of
the Deltoidal Hexecontahedron or vertices of its dual,
the Rhombicosidodecahedron. Since the graph of a
Rhombicosidodecahedron (Figure 1) allows the study
of each protein as a vertex, in what follows we mainly
used that perspective to develop the following meth-
ods of the analysis of virus disassembly.
2.2 Structures of Viruses’ Capsids
Capsids’ atomic coordinates were obtained from the
Protein Data Bank (PDB, http://www.rcsb.org/,
(Berman et al., 2000)). Only T = 1 cap-
sids, present on ViperDB were considered
(http://viperdb.scripps.edu, (Carrillo-Tripp et al.,
2009)). The capsid structures analysed in this work
were divided into groups according to the criteria of
similarity of the infection host and the genes codify-
ing capsid proteins. The list is presented in Table 2,
with 51 structures divided into 14 groups. Human
Adenovirus are mainly T = 25 and the structures
studied here are a stable capsid formed with the
pentagons of the T = 25, forming a Dodecahedron
(named from now on as Human Adenovirus Pt-Dd or
HAPD).
2.3 Energy Calculation
To calculate the energy of the full capsid and its dis-
assembly products, the number of hydrogen bonds,
salt bridges and hydrophobic contacts between cap-
sid subunits were counted (intra-protein contacts were
ignored). Hydrogen bonds (N
HB
) were counted if ac-
ceptor and donor’s atoms indicated in Table 1 were
BIOINFORMATICS 2017 - 8th International Conference on Bioinformatics Models, Methods and Algorithms
218

Table 1: Donor and Acceptor’s Atoms used to calculate Hy-
drogen Bonds.
Amino acids
Donor
ARG(N
ε
; N
η1
; N
η2
), ASN(N
δ2
), CYS(S
γ
),
GLN(N
ε2
), HIS(N
δ1
; N
ε2
), LYS(N
ζ
),
SER(O
γ
), THR(O
γ1
), TRP(N
ε1
), TYR(O
η
)
Acceptor
ASN(O
δ1
), ASP(O
δ1
; O
δ2
), GLN(O
ε1
),
GLU(O
ε1
; O
ε2
), HIS(N
δ1
; N
ε2
),
SER(O
γ
), THR(O
γ1
), TYR(O
η
)
at a distance less than or equal to 4.0
˚
A. A salt bridge
bound (N
SB
) was counted if a positively charged atom
of an acidic amino acid (ASP and GLU) was found
within 4.0
˚
A of a negatively charged atom of a basic
amino acid (ARG and LYS). A hydrophobic contact
(N
HC
) was counted when β-carbons of the residues
ALA, VAL, LEU, ILE, MET, PHE, TYR and TRP,
were found at the distance less than or equal to 7.0
˚
A.
We can calculate a heuristic measure of total energy
of a complete capsid or capsid fragment (Equations
(1) to (3)).
E
1
= N
SB
+ N
HB
+ N
HC
(1)
E
2
= 20 × N
SB
+ N
HB
+ N
HC
(2)
E
3
= 100 × N
SB
+ 10 × N
HB
+ N
HC
(3)
In Heuristic I (Equation (1)), only the total num-
ber of inter-subunit bonds in the capsid complex is
taken into account; the three types of bounds are
equally weighted. Heuristic II (Equation (2)) gives
Salt Bridges 20 times more energy than Hydrogen
Bonds. Hydrophobic Contacts was considered to be
energetically equivalent to making Hydrogen Bonds,
since a water ”cage” is destroyed when hiding hy-
drophobic amino acids, breaking Hydrogen Bonds
made by the water itself to hold this cage (Atkins and
De Paula, 2006). On the other hand, on Heuristic III
(Equation (3)) an increasing power of 10 was given to
each type of inter-subunit bond, in the known order of
strength of this types of contacts (Salt Bridges > Hy-
drogen Bonds > Hydrophobic Contacts) (Atkins and
De Paula, 2006).
2.4 Removal of Proteins and Graph
Representation
Each possible capsid fragment can be represented by
a 60 element binary vector, depending on the presence
or absence of each protein given the subunit number-
ing of Figure 1. A list of non-redundant binary vectors
representing capsid fragments was then obtained as
follows: for every combination of N indexes of a size
60 vector, a binary vector was generated with zeros
in those positions and ones elsewhere; permutations
of the permutation group mentioned above were ap-
plied to this vector; if any of the resulting vectors was
not identical to another previously obtained it would
be appended to the list. Thus, a set of non geometri-
cally identical capsid fragments was obtained. Their
representation as a subgraph of Figure 1 allowed us
to check if disconnected fragments were obtained at
each step, so that the largest fragment was retained
and the search for non redundant forms was also ap-
plied to this fragment. Finally, the fragment’s the en-
ergy was calculated according to each heuristic.
2.5 Optimal Path for Disassembly
For all the possibilities of removing N proteins, a tree
of all the paths that the virus disassembly process
could take was built, rooted at the intact virus cap-
sid (N = 0). Edges were given a transition energy
weight, based on the average heuritic energy per pro-
tein, following Equation 4. The size of configuration
i is equal to 60− N
i
, where N
i
was the number of pro-
teins removed.
E
transition i→ j
=
E
j
size
j
−
E
i
size
i
(4)
The Bellman-Ford algorithm was used on this tree
to calculate the shortest path from the complete vi-
ral capsid to every possibility of a capsid with 60 − N
proteins, based on the weights of the edges of the tree,
recording the five shortest paths for every N and the
corresponding final configuration.
2.6 Implementation
All methods were implemented in Python 2.7
using scientific computing modules (igraph
Python bindings (Csardi and Nepusz, 2006)).
All the code is available on GitHub repository:
https://github.com/CAPiedade/Virus-Disassembly.
3 RESULTS AND DISCUSSION
The disassembly paths of the different capsids are
portrayed in Table 2 considering heuristic I. It can
be observed that most Parvoviruses (marked with
a †) follow the same sequence of disassembly:
{1} → {1,10} → {1, 10, 23} → {1, 2,10,23} →
{1,2,10,22,23}. Fragment {1, 10, 23} represents
the removal of a triangle of proteins on the capsid
structure (Figure 2.A). As observed in the mechanical
removal of proteins with AFM (Castellanos et al.,
2012), the Minute Mice Virus (on our work repre-
sented by the group of the Rodent Protoparvovirus)
How to Disassemble a Virus Capsid - A Computational Approach
219
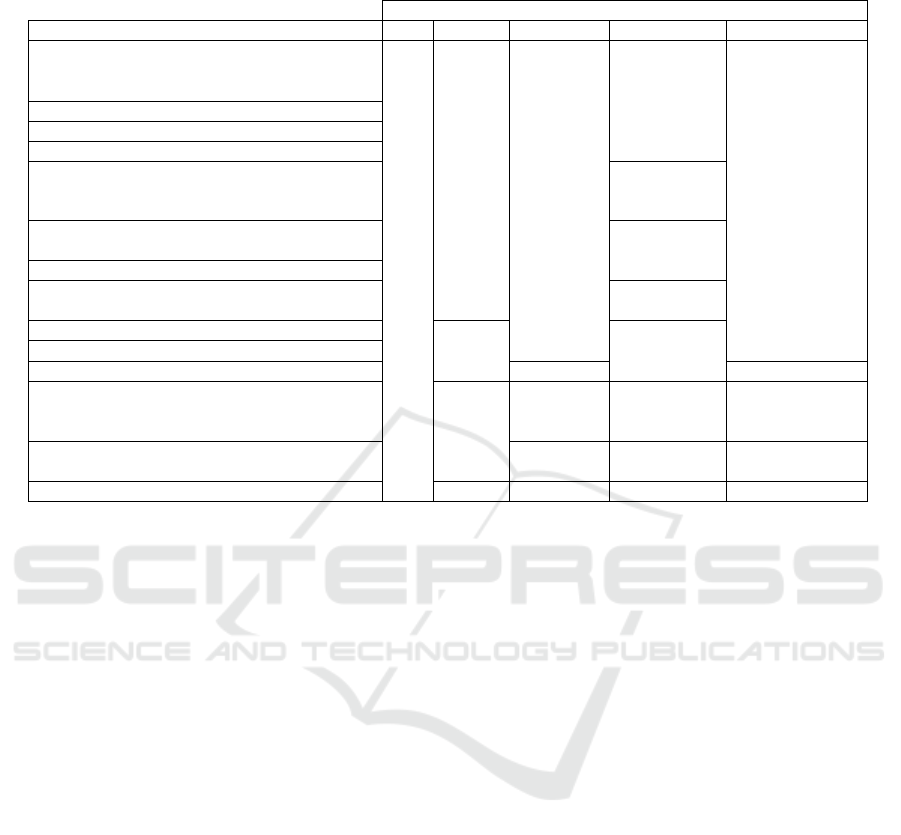
Table 2: Capsid groups and minimal energy paths taken to reach the removal of N subunits using heuristic I. Viruses marked
with † are Parvoviruses (Family Parvoviridae).
N
Groups 1 2 3 4 5
Adeno-Associated Virus†
(3j4p, 3ra8, 4iov, 3j1q, 3ntt, 3ra2, 3ux1,
2g8g, 1lp3, 2qa0, 3ra4, 3ra9, 3raa, 4rso, 5egc)
{1}
{1,10}
{1,10,23}
{1,2,10,23}
{1,2,10,22,23}
Bovine Parvovirus† (4qc8)
Human Parvovirus† (1s58)
Porcine Parvovirus† (1k3v)
Canine and Feline Panleukopenia Virus†
(1c8f, 1c8h, 1c8e, 1c8g, 1fpv, 1p5y, 1c8d,
1p5w, 4dpv, 1ijs, 2cas)
{1,9,24,54}
{1,2,10,23}
Rodent Protoparvovirus†
(1mvm, 1z14, 4g0r, 1z1c, 2xgk, 4gbt)
{1,2,10,23}
Avian Birnavirus (1cwd)
Porcine Circovirus (3r0r, 3jci)
{1,2,22,42}
{1,2,10,23}
Bombyx mori Densovirus† (3p0s)
{1,2} {1,2,22,23}Galleria mellonella Densovirus† (1dnv)
Penaeus stylirostris Densovirus† (3n7x)
{1,2,23} {1,2,6,10,23}
Satellite Tobacco Mosaic Virus
(2buk, 4bcu, 1a34, 4oq8)
{1,6}
{1,2,6}
{1,2,23}
{1,2,6,7}
{1,2,6,24}
{1,2,6,23}
{1,2,6,10,24}
{1,2,3,23,42}
{1,2,6,10,23}
Hepatitis E Virus (HEV) (2ztn, 2zzq, 3hag)
{1,2,23}
{1,2,10,23}
{1,2,6,23}
{1,2,6,10,23}
Human Adenovirus Pt-Dd (1x9t, 4aqq, 4ar2)
{1,2} {1,2,3} {1,2,3,4} {1,2,3,4,5}
tends to start the disassembly process by losing
a triangular block, which is then followed by the
removal of the adjacent triangle. This is supported
by our results since most disassembly pathways lead
to the removal of a triangular structure, commonly
composed of proteins {1, 10,23}. The theoretical
studies of Rapaport et al. (Rapaport, 2008; Rapaport,
2010) reveal the existence of long-lived transient
structures with just one last triangle of proteins
missing to form the complete capsid. The results
of Reddy et al. (Reddy et al., 1998) using both
individual proteins and trimers show the same
for capsid assembly (Reddy et al., 1998), sug-
gesting that the path for disassembly also follows
the triangle removal. Fragment {1,2,10,22,23}
represents the removal of a trapezium-like shape,
centred on the triangle {1,10,23} (Figure 2.C)
Supposing this trend continues, it is not hard to
see that there is a chance of removing the proteins
around the five-fold axis. These could be the fol-
low up steps of the disassembly of these capsids,
potentially resulting in the loss of 15 proteins such
as {1,2,3,4, 5, 6, 10,22,23,33,34, 41, 42,49,50}.
Castellanos et al.’s results confirms the predictions of
Reddy et al. (Reddy et al., 1998), since the lowest
energy configuration, just before the complete capsid,
was missing a pentamer of triangles (corresponding
to the 15-protein combinations described before),
inclusively for Parvoviruses (Reddy and Johnson,
2005). The removal of four proteins has more than
one possibility. Most parvoviruses lose subunits
{1,2,10,23} (Figure 2.B). Densovirus, on the other
hand, tend to lose proteins {1,2,22,23}, a config-
uration leading to a square hole on the two-fold
symmetry of the capsid (Figure 2.D). We might
speculate that the inter-subunit interactions on these
viruses are different from the others parvoviruses.
To verify this, we took the ratio between the number
of intersubunit contacts [22 ↔ 23] and the number
of intersubunit contacts [10 ↔ 23], the only two
interactions between capsid proteins that are different
between {1,2, 22, 23} and the common parvovirus
structure {1,2,10,23}, respectively. These results
are shown on Table 3, where it can be observed that
Parvoviruses have a lower ratio than Densovirus,
suggesting that
#[22↔23]
/
#[10↔23]
> 0.82 ⇒ {1,2,22,23}
#[22↔23]
/
#[10↔23]
< 0.80 ⇒ {1,2,10,23}
Further analysis of these and other ratios will provide
some insight into why some structures are oriented
into particular pathways. Removing of a square-
shaped group of proteins raises the idea that the higher
the symmetry of the structures removed, the higher is
their chance to be lost. Structures with PDB
ID
1c8f
and 1c8h differ only inside its group for the removal
of the four proteins {1, 9, 24, 54}. Avian Birnavirus
follow the same path as the majority of Parvovirus,
as well as Porcine Circovirus, with the exception of
structure with PDB
ID
3jci which loses {1,2,22,42},
BIOINFORMATICS 2017 - 8th International Conference on Bioinformatics Models, Methods and Algorithms
220

Figure 2: Disassembly stages of different viruses. Parvoviruses (PDB
ID
1s58) path {1,10,23} (A), {1,2,10,23} (B) and
{1,2,10,22,23} (C). Densovirus (PDB
ID
1dnv) path {1,2,22,23} (D). HEV (PDB
ID
2zzq) path {1,2,6,10,23} (E). HAPD
(PDB
ID
4aqq) path {1,2,3,4,5} (F). Structures are represented showing molecular surfaces. UCSF Chimera (Pettersen et al.,
2004) was used for rendering.
Table 3: Ratio of contacts between proteins #[22 ↔ 23] and #[10 ↔ 23].
Ratio of the number
of contacts
Densovirus Parvovirus
1dnv 3n7x 3p0s 1s58 3raa
#[22↔23]
/
#[10↔23]
18
/
19
≈ 0,95
15
/
11
≈ 1,36
23
/
28
≈ 0,82
35
/
44
≈ 0,80
35
/
62
≈ 0,56
a reflection of {1, 2, 10,23}. HEV capsids loses
proteins on the order {1} → {1, 6} → {1, 2,23} ⇒
{1,2,6,23}
{1,2,10,23}
⇒ {1,2,6,10,23}. Although distinguish-
able from the Parvovirus, HEV loses a pentamer that
forms a triangular hole on the capsid structure (Fig-
ure 2.E). HAPD follows the disassembly path {1} →
{1,2} → {1,2,3} → {1, 2, 3,4} → {1,2,3,4, 5}, an
exception to all the others. The Human Adenovirus
group is the only undergoing a five-fold removal
of the proteins, removing the pentamer {1,2,3,4,5}
(Figure 2.F). On Figure 2.F we can observe the struc-
ture of a HAPD, which is formed by very condensed
clusters of pentagons, having very few contacts with
the 2-fold and 3-fold proteins. The distance between
the pentagonal clusters on the full capsid structure
might make it easier for this set of subunits to be re-
moved, in opposition to creating a bigger gap by re-
moving, for example, proteins {1,2, 10, 22,23}. Our
results are supported by those of Ortega-Esteban et al.
(Ortega-Esteban et al., 2013) which showed, through
AFM, a loss of a pentagon-shaped pentamer of pro-
teins for Human Adenoviruses. Satellite Mosaic To-
bacco Virus disassembly was to diverse. The results
from this group were not analysed any further since
due to the small size of the set.
We also investigated the effect of the other two
heuristics (Equations 2 and 3). Results were very
similar to heuristic I, with a slightly higher degree
of branching. Nevertheless, the loss of three and five
proteins was conserved in all heuristics.
4 CONCLUSIONS
Our study began with the goal of investigating
whether there was a common disassembly pathway
among different virus families and, if not, if there
would be a conserved disassembly path for each fam-
ily. Results on this work support the idea that, for a
large cluster of T = 1 viruses, there is a common dis-
assembly pathway. This cluster is composed by some
Parvoviruses (Adeno-Associated Virus, Bovine Par-
vovirus, Human Parvovirus, Porcine Parvovirus, Ro-
dent Protoparvovirus, Canine and Feline Panleukope-
nia Virus), Avian Birnavirus, Hepatitis E Virus and
Porcine Circovirus. From our results, we can specu-
late that the results of Castellanos et al. (Castellanos
et al., 2012) might be applicable to more families of
viruses besides the Rodent Protoparvovirus. Denso-
viruses (from Bombyx mori, Galleria mellonella and
Penaeus stylirostris) are exceptional for their ex-
clusive removal of proteins {1,2,22,23}, with no
branching to alternative possibilities. On the other
hand, Human Adenoviruses have a very particular
pathway of disassembly, establishing that not all dis-
assemblies proceed through the removal of triangular
protein trimers, but also through the removal of pro-
tein pentagons. Using a combinatorial method based
on symmetry and geometry, with a 60-subunit model
(Deltoidal Hexecontahedron) and not a 20-subunit
model (Icosahedron) or 12-subunit model (Dodeca-
hedron), under a heuristic using the number of weak
How to Disassemble a Virus Capsid - A Computational Approach
221

contacts, we obtained results comparable to those
found in previous literature. Furthermore, the usage
of a 60-subunit model allows the study of cases such
as those of Human Adenoviruses, which do not lose
triangular faces of the Icosahedron, but a pentagon
of faces of the Deltoidal Hexecontahedron. Nonethe-
less, an increase of the sample size is an important
follow-up step, since it would give a better insight into
the trends observed for the different viruses studies.
Moreover, the number of subunits removed from the
capsid structures should be above 5 to offer a perspec-
tive of what would be the next steps on the disassem-
bly pathway. Improvement of the algorithms as well
as the processing power could provide more informa-
tive results beyond 5. Understanding the way viruses’
capsids disassemble can be used to interfere with the
inter-subunit interactions, specially those who hold
the triangular trimer in the complete capsid. Antiviral
drugs can thus be design to disrupt these interactions.
ACKNOWLEDGEMENTS
Work supported by project RECI/BBB-
BEP/0104/2012 from Fundac¸
˜
ao para a Ci
ˆ
encia
e Tecnologia, Portugal. The funders had no role in
study design, data collection and analysis, decision to
publish, or preparation of the manuscript.
REFERENCES
Atkins, P. W. and De Paula, J. (2006). Physical chemistry
for the life sciences. Oxford University Press ; W.H.
Freeman, Oxford, UK : New York.
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat,
T. N., Weissig, H., Shindyalov, I. N., and Bourne, P. E.
(2000). The protein data bank. Nucleic acids research,
28(1):235–242.
Burnside, W. (1909). Theory of groups of finite order. Mes-
senger of Mathematics, 23:112.
Carrillo-Tripp, M., Shepherd, C. M., Borelli, I. A.,
Venkataraman, S., Lander, G., Natarajan, P., John-
son, J. E., Brooks, C. L., and Reddy, V. S. (2009).
Viperdb2: an enhanced and web api enabled rela-
tional database for structural virology. Nucleic acids
research, 37(suppl 1):D436–D442.
Caspar, D. L. D. and Klug, A. (1962). Physical Principles
in the Construction of Regular Viruses. Cold Spring
Harbor Symposia on Quantitative Biology, 27(0):1–
24.
Castellanos, M., Prez, R., Carrillo, P., dePablo, P., and
Mateu, M. (2012). Mechanical Disassembly of
Single Virus Particles Reveals Kinetic Intermedi-
ates Predicted by Theory. Biophysical Journal,
102(11):2615–2624.
Csardi, G. and Nepusz, T. (2006). The igraph software
package for complex network research. InterJournal,
Complex Systems:1695.
Horton, N. and Lewis, M. (1992). Calculation of the free
energy of association for protein complexes. Protein
Science, 1(1):169–181.
Mateu, M. G. (2013). Assembly, stability and dynamics
of virus capsids. Archives of Biochemistry and Bio-
physics, 531(1-2):65–79.
Ortega-Esteban, A., Prez-Bern, A. J., Menndez-Conejero,
R., Flint, S. J., Martn, C. S., and de Pablo, P. J. (2013).
Monitoring dynamics of human adenovirus disassem-
bly induced by mechanical fatigue. Scientific Reports,
3.
Perlmutter, J. D. and Hagan, M. F. (2015). Mechanisms of
Virus Assembly. Annual Review of Physical Chem-
istry, 66(1):217–239.
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S.,
Greenblatt, D. M., Meng, E. C., and Ferrin, T. E.
(2004). Ucsf chimeraa visualization system for ex-
ploratory research and analysis. Journal of computa-
tional chemistry, 25(13):1605–1612.
Poranen, M. M., Daugelaviius, R., and Bamford, D. H.
(2002). Common Principles in Viral Entry. Annual
Review of Microbiology, 56(1):521–538.
Prasad, B. V. V. and Schmid, M. F. (2012). Principles of
Virus Structural Organization. In Rossmann, M. G.
and Rao, V. B., editors, Viral Molecular Machines,
volume 726, pages 17–47. Springer US, Boston, MA.
Rapaport, D. C. (2008). Role of Reversibility in Viral Cap-
sid Growth: A Paradigm for Self-Assembly. Physical
Review Letters, 101(18).
Rapaport, D. C. (2010). Studies of reversible capsid shell
growth. Journal of Physics: Condensed Matter,
22(10):104115.
Reddy, V. S., Giesing, H. A., Morton, R. T., Kumar,
A., Post, C. B., Brooks, C. L., and Johnson, J. E.
(1998). Energetics of Quasiequivalence: Compu-
tational Analysis of Protein-Protein Interactions in
Icosahedral Viruses. Biophysical Journal, 74(1):546–
558.
Reddy, V. S. and Johnson, J. E. (2005). Structure-Derived
Insights into Virus Assembly. In Advances in Virus
Research, volume 64, pages 45–68. Elsevier.
Vincent, A. (2001). Molecular symmetry and group theory:
a programmed introduction to chemical applications.
Wiley, Chichester ; New York, 2nd ed edition.
Zlotnick, A. (1994). To build a virus capsid. An equilib-
rium model of the self assembly of polyhedral protein
complexes. J. Mol. Biol., 241(1):59–67.
Zlotnick, A. (2003). Are weak proteinprotein interac-
tions the general rule in capsid assembly? Virology,
315(2):269–274.
Zlotnick, A., Johnson, J. M., Wingfield, P. W., Stahl, S. J.,
and Endres, D. (1999). A theoretical model success-
fully identifies features of hepatitis B virus capsid as-
sembly. Biochemistry, 38(44):14644–14652.
Zlotnick, A. and Stray, S. J. (2003). How does your virus
grow? Understanding and interfering with virus as-
sembly. Trends Biotechnol., 21(12):536–542.
BIOINFORMATICS 2017 - 8th International Conference on Bioinformatics Models, Methods and Algorithms
222
