
Enhancing Emotion Recognition from ECG Signals using Supervised
Dimensionality Reduction
Hany Ferdinando
1,2
, Tapio Seppänen
3
and Esko Alasaarela
1
1
Optoelectronics and Measurement Technique Research Unit, University of Oulu, Oulu, Finland
2
Department of Electrical Engineering, Petra Christian University, Surabaya, Indonesia
3
Physiological Signal Analysis Team, University of Oulu, Oulu, Finland
{hany.ferdinando, tapio, esko.alasaarela}@ee.oulu.fi
Keywords: Emotion Recognition, kNN, Dimensionality Reduction, LDA, NCA, MCML.
Abstract: Dimensionality reduction (DR) is an important issue in classification and pattern recognition process. Using
features with lower dimensionality helps the machine learning algorithms work more efficient. Besides, it
also can improve the performance of the system. This paper explores supervised dimensionality reduction,
LDA (Linear Discriminant Analysis), NCA (Neighbourhood Components Analysis), and MCML (Maximally
Collapsing Metric Learning), in emotion recognition based on ECG signals from the Mahnob-HCI database.
It is a 3-class problem of valence and arousal. Features for kNN (k-nearest neighbour) are based on statistical
distribution of dominant frequencies after applying a bivariate empirical mode decomposition. The results
were validated using 10-fold cross and LOSO (leave-one-subject-out) validations. Among LDA, NCA, and
MCML, the NCA outperformed the other methods. The experiments showed that the accuracy for valence
was improved from 55.8% to 64.1%, and for arousal from 59.7% to 66.1% using 10-fold cross validation after
transforming the features with projection matrices from NCA. For LOSO validation, there is no significant
improvement for valence while the improvement for arousal is significant, i.e. from 58.7% to 69.6%.
1 INTRODUCTION
Decreasing the dimensionality of features without
losing their important characteristic is a vital pre-
processing phase in high-dimensional data analysis
(Sugiyama, 2007). Dimensionality reduction (DR) is
an important tool to handle the curse of
dimensionality. Projecting high dimensional feature
space to lower dimensional feature space helps
classifiers perform better. As human vision system is
limited to 3D, visualization of feature space gets
benefits from DR. Moreover, DR is also useful in data
compression (Lee and Verleysen, 2010), for example
when it is important to store all training data as in k-
nearest neighbour classifier (kNN).
Dimensionality reduction (DR) methods include
linear and nonlinear techniques. Well known method
for linear DR is principal component analysis (PCA)
(Jolliffe, 2002). The nonlinear DR emerged later, e.g.
Sammon’s mapping (Sammon, 1969). Furthermore,
there are supervised and unsupervised DR techniques.
The supervised DRs use labels of the data to guide the
mapping process while the unsupervised ones rely on
finding a projection space which provides the highest
variance.
This paper explores a number of supervised DR
techniques, i.e. Neighbourhood Components
Analysis (NCA), Linear Discriminant Analysis
(LDA), Maximally Collapsing Metric Learning
(MCML), and applied them to enhance the accuracy
of emotion recognition-based ECG signal from the
Mahnob-HCI database for affect recognition.
The Mahnob-HCI database was published in 2012
with some baseline accuracies (Soleymani, et al.,
2012) for 3-class classification problem of valence
and arousal. However, a baseline for emotion
recognition based on ECG signals only were not
given therein. Ferdinando et al. (Ferdinando, et al.,
2014) computed Heart Rate Variability (HRV)
indexes achieving baseline accuracies, 42.6% and
47.7% for valence and arousal respectively. Later,
Ferdinando et al. improved the accuracy to 55.8% and
59.7% for valence and arousal respectively by
applying bivariate empirical mode decomposition
(BEMD) to ECG signals and use the statistical
distributions of dominant frequency as the features
(Ferdinando, et al., 2016).
112
Ferdinando, H., Seppänen, T. and Alasaarela, E.
Enhancing Emotion Recognition from ECG Signals using Supervised Dimensionality Reduction.
DOI: 10.5220/0006147801120118
In Proceedings of the 6th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2017), pages 112-118
ISBN: 978-989-758-222-6
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

Although significant improvements have been
achieved in (Ferdinando, et al., 2016), the best
accuracies, so far, from this database were 76% and
68% for valence and arousal respectively (Soleymani,
et al., 2012) using features from eye gaze and EEG.
We aim at improving the classification accuracy by
using only ECG signals.
In this paper, we enhance the accuracy of emotion
recognition by applying supervised DR to the features
based on applying BEMD analysis to ECG signals
(Ferdinando, et al., 2016) prior feeding them to the
kNN classifier. Projection matrix calculations were
done with the Matlab code by van der Maaten (van
der Maaten, 2016).
2 SUPERVISED
DIMENSIONALITY
REDUCTION
Supervised DRs in drtoolbox are Linear Discriminant
Analysis (LDA), Generalized Discriminant Analysis
(GDA), Neighbourhood Components Analysis
(NCA), Maximally Collapsing Metric Learning
(MCML), and Large Margin Nearest Neighbor
(LMNN) (van der Maaten, 2016). They work based
on the label/class of the inputs. The labels serve as a
guideline to reduce the dimensionality. The
supervised DR methods in this exploration are based
on a Mahalonobis distance measure
2121
2
21
xxAxxxx
T
ff
(1)
within kNN framework, except LDA and GDA,
where
WWA
T
is a positive semidefinite (PSD)
matrix, and W is the projection matrix to a certain
space. The ultimate goal is to find projection matrix
A, such that the classifiers perform well in the
transformed space. Unfortunately, the GDA does not
provide a projection matrix A such that new features
can be transformed into other space but user can
choose the target dimensionality (van der Maaten,
2016). For this reason, GDA was not included in our
study. Looking to the implementation of LMNN,
there is no such dimensionality reduction but it
provides a projection matrix A (van der Maaten,
2016). Due to this fact, the LMNN was also discarded
from the experiments.
2.1 Linear Discriminant Analysis
(LDA)
Linear Discriminant Analysis (LDA) (Weinberger
and Saul, 2009) computes linear projection
ii
xx A
that maximizes the amount of between-class variance
(C
b
) relative to the amount of within-class variance
(C
w
). The objective function is defined as
ACA
ACA
w
T
b
T
TraceAf )(
subject to
IAA
T
(2)
The LDA DR works well when the reduced
dimensionality is less than the number of classes. In
addition, the conditional densities of the classes must
be multivariate Gaussian. Failing to fulfil this
requirement makes the transformed features not
suitable for kNN. This method has been applied to
spoken emotion recognition problem (Zhang and
Zhao, 2013), EEG-based emotion recognition
(Valenzi, et al., 2014), and ECG-based individual
identification (Fratini, et al., 2015).
2.2 Neighbourhood Components
Analysis (NCA)
Neighbourhood Component Analysis (NCA)
(Goldberger, et al., 2005) is non-parametric which
makes no assumption about the shape of the class
distribution or the boundaries between them. The
algorithm directly maximizes a stochastic variant of
the leave-one-out kNN score on the training set. The
final goal is to find a transformation matrix such that
in the transformed space, the kNN performs well. The
size of the transformation matrix determines the
dimension of the transformed features. Using this
method, one can visualize high dimensional features
in 2D or 3D space.
To deal with the discontinuity of the leave-one-
out classification error of kNN, a differentiable cost
function based on stochastic (“soft”) neighbour
assignment in the transformed space was introduced
(Goldberger, et al., 2005). The idea is to use softmax
function, to transform distance from point i to j into
probability p
ij
and inherit its class label from the
selected point.
ik
ki
ji
ij
p
2
2
exp
exp
AxAx
AxAx
,
0
ii
p
(3)
with objective function defined as
Enhancing Emotion Recognition from ECG Signals using Supervised Dimensionality Reduction
113

i
i
iCj
ij
ppAf
i
)(
(4)
The algorithm searches for the transformation matrix
A, such that the objective function is maximized. The
algorithm uses a gradient rule, by differentiating f(A)
with respect to the transformation matrix A, for
learning. The NCA was able to separate data
containing useful information and noise, which ended
up with dimensionality reduction (Goldberger, et al.,
2005).
The NCA has been applied to research in
Affective Computing, e.g. Zhang and Zhao applied it
to the spontaneous Chinese and the acted Berlin
database for spoken emotion recognition and then
compared it with other dimensionality reduction
methods (Zhang and Zhao, 2013). McDuff et al. used
the NCA in AffectAura project (McDuff, et al.,
2012). Romero et al. put on the NCA to reduce
dimensionality of features from EEG (Romero, et al.,
2015).
2.3 Maximally Collapsing Metric
Learning (MCML)
Maximally Collapsing Metric Learning (MCML)
(Globerson and Roweis, 2006) uses simple geometric
intuition that all points belonging to the same class
are mapped (collapsed) to a single location in feature
space and all points from the other classes are mapped
to other locations. The main goal is to find a
transformation matrix A such that it fulfills the simple
geometric intuition idea. To learn the distance
measure, each training point is assigned to a
conditional probability,
A
ij
A
pijp |
, over other
points using softmax function. From conditional
probability point of view, the probability of a point
belonging to class X given that point is in class X is
1, otherwise it is zero. Given pairs of input and label
ii
yx ,
, the conditional probability is defined as
ij
ij
ij
yy
yy
p
,0
,1
*
(5)
The algorithm searches for a matrix A such that
A
ij
p
is as close as possible to
*
ij
p
by minimizing
objective function f(A), i.e. Kullback-Leibler
divergence between them, such that
PSDA
. The
objective function (Globerson and Roweis, 2006) is
defined as
i
A
ijpijpKLAf |||)(
0
(6)
The MCML has been applied to spoken emotion
recognition (Zhang and Zhao, 2013) and EEG-based
Iyashi expression analysis (Romero, et al., 2015).
3 MATERIAL AND METHODS
3.1 ECG Signal Processing
The Mahnob-HCI database contains 32-channel
EEG, peripheral physiological signals (ECG,
temperature, respiration, skin conductance), face and
body video, speech, and eye gaze recording from 27
subjects (11 males and 16 females). All signals were
precisely synchronized which is suitable for
multimodal emotional response studies. The ECG
signals were sampled at 256 Hz (Soleymani, et al.,
2012).
We used the same data as in (Ferdinando, et al.,
2014), i.e. “Selection of Emotion Elicitation” in the
database. The original data contains 513 samples.
However, the sample from session 2508 was
discarded because visual inspection showed it is
corrupted. Thus, we worked with 512 samples,
subject to several filters to suppress noise from power
line interference, baseline drift, motion artifact,
electrode contact, and muscle contraction
(Soleymani, et al., 2012).
The ECG signals contain data from both
unstimulated and stimulated phase. Since we were
only interested in ECG during stimulated phase, this
part must be separated from the other utilizing
synchronization signal provided by the database.
The BEMD method (Rilling, et al., 2007) was
used to get features from ECG. Based on our
experiments, the BEMD method was sensitive to the
length of the signal. For this reason, the ECG signal
was divided into 5 second segments. A synthetic ECG
signal, synchronized with the R-wave event to the
original signal, was generated by using the model
from McSharry et al. (McSharry, et al., 2003). This
signal served as the imaginary part of the ECG signal
while the original served as the real part. This
complex-valued ECG signal was analyzed by the
BEMD method, resulting in 5-6 intrinsic mode
functions (IMFs). The first three IMFs, as suggested
by Agrafioti et al. (Agrafioti, et al., 2012), were
analyzed for dominant frequencies using spectrogram
analysis (Ferdinando, et al., 2016). The spectrogram
analysis relies on two parameters, i.e. window size
ICPRAM 2017 - 6th International Conference on Pattern Recognition Applications and Methods
114

and overlap. The dominant frequencies of all 5 second
segments are collected and various features are
calculated as follows. The features are based on the
statistical distribution of the dominant frequencies
and their first difference: mean, standard deviation,
median, Q1, Q3, IQR, skewness, kurtosis, percentile
2.5, percentile 10, percentile 90, percentile 97.5,
maximum, and minimum. The results are groups in
three sets: feature1 (statistical distribution of the
dominant frequencies; 84 features), feature2
(statistical distribution of the dominant frequencies’
first difference; 84 features), and feature12 (combine
both feature1 and feature2; 168 features). The best
features are then selected from each group with
sequential forward-floating search. The number of
most selected features varies from two to twenty-
three, depending on whether valence or arousal is
recognized and the parameters used in the
spectrogram analysis (Ferdinando, et al., 2016).
3.2 Dimensionality Reduction
The chosen DR methods, LDA, NCA, and MCML,
are applied to the selected features from certain
window size and overlap parameters combination in
the spectrogram analysis found in (Ferdinando, et al.,
2016) to get features with lower dimensionality. The
initial matrix
A for NCA and MCML are generated
with a random number generator. It means that there
is no guarantee that they provide the optimum matrix
A in one pass. The algorithm is modified to be
iterative such that it stops – a flag is set – when there
is no improvement, validated using leave-one-out
cross-validation, within 200 iterations. The DR is
applied only in cases when the number of selected
features is greater than the target dimensionality. The
optimum projection matrix
A is saved for further
process.
3.3 Classifier and Validation Methods
We used kNN classifier as in (Ferdinando, et al.,
2016) to solve the original 3-class classification
problem for valence and arousal. 20% of the data are
held out for validation while the rest are subject to 10-
fold cross validation. The classifier model is built
based on the projection of the selected features using
the optimum projection matrix
A during the DR
phase. The whole validation process is repeated 100
times with new resampling in each iteration. The
average over the repetition represent the final
accuracy. When the accuracies from different
combinations of window size and overlap parameter
are close to each other, the final accuracy is justified
using the Law of Large Numbers (LLN).
Another validation for the result is leave-one-
subject-out (LOSO) validation. The main idea is to
evaluate if the transformed features are general
enough to work well with features from new subjects.
4 RESULTS
Table 1 to 4 show the best results from each target
dimensionality of each DR algorithm with 10-fold
cross validation and 100 iterations.
Table 1: Accuracy after applying LDA DR for valence and
arousal.
Dimensionality Valence Arousal
2D 55.1 ± 7.4
59.9 ± 6.8
Since this is 3-class problem, the highest
dimensionality that the LDA can yield is two.
Surprisingly, the accuracy for both valence and
arousal are close to (Ferdinando, et al., 2016). An
improvement, however, is less storage and faster
calculation than standard kNN.
Table 2: Accuracy after applying NCA DR for valence and
arousal.
Dimensionality Valence Arousal
2D 61.3 ± 7.2 65.6 ± 6.2
3D 57.0 ± 8.0 66.0 ± 8.1
4D 65.3 ± 6.5 60.1 ± 7.7
5D 64.5 ± 6.7 61.0 ± 8.1
6D 53.2 ± 7.6 61.5 ± 7.5
7D 60.4 ± 6.6 61.2 ± 7.2
Results from the NCA for both valence and
arousal are promising, since the best accuracies for
valence and arousal are even higher than in
(Ferdinando, et al., 2016).
Table 3: Accuracy after applying MCML DR for valence
and arousal.
Dimensionality Valence Arousal
2D 54.5 ± 7.9 60.5 ± 7.5
3D 54.6 ± 7.4 48.9 ± 7.3
4D 41.8 ± 6.9 49.3 ± 7.2
5D 41.9 ± 7.2 49.3 ± 7.1
6D 42.1 ± 7.6 49.2 ± 7.0
7D 43.5 ± 7.3 48.4 ± 8.9
The best results based on the MCML DR from
both valence and arousal are close to the ones in
(Ferdinando, et al., 2016). It also results in less
Enhancing Emotion Recognition from ECG Signals using Supervised Dimensionality Reduction
115
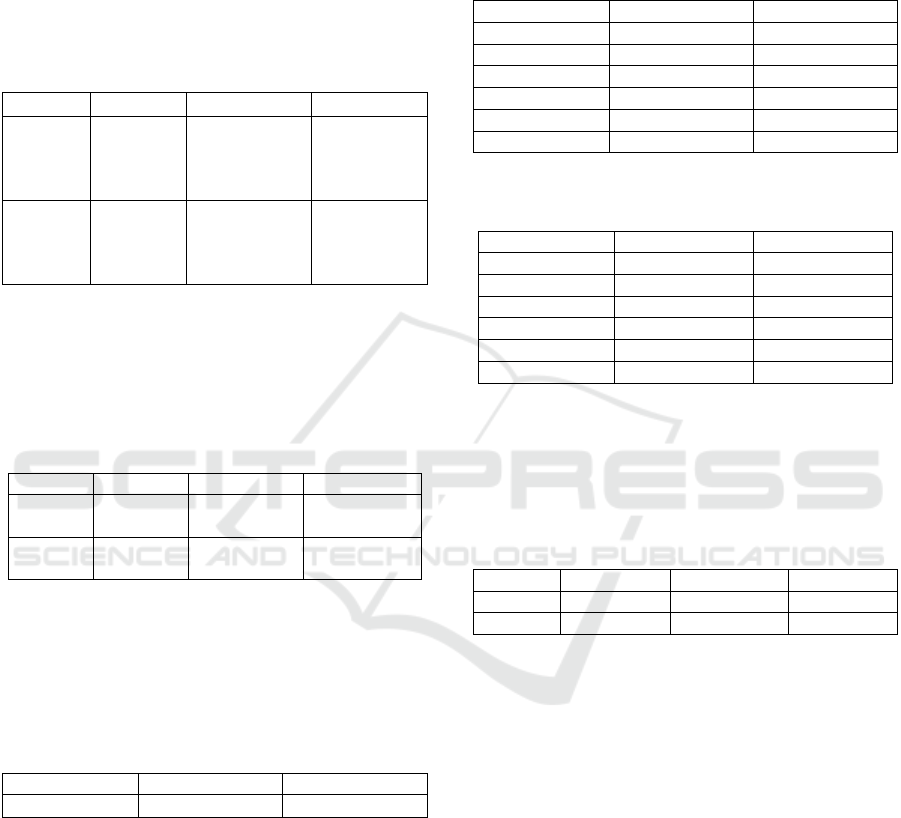
storage for the data and faster computation than
standard kNN.
Table 4 compares the results among LDA, NCA,
and MCML side-by-side. It shows that the NCA
outperforms the other methods. The difference is
roughly 10% and 5% for valence and arousal,
respectively.
Table 4: Best accuracies of the dimensionality reduction
methods.
LDA NCA MCML
Valence
55.1 ± 7.4 65.3 ± 6.5
(4D)
64.5 ± 6.7
(5D)
54.6 ± 7.4
(3D)
54.5 ± 7.9
(2D)
Arousal
59.9 ± 6.8 66.0 ± 8.1
(3D)
65.6 ± 6.2
(2D)
60.5 ± 7.5
(2D)
Since the most promising results in some cells in
Table 4 are close to each other, the Law of Large
Numbers is used to estimate accuracies as close as
possible to the true ones. After 1000 iterations, the
best results are in Table 5.
Table 5: Applying LLN based on Table 4.
LDA NCA MCML
Valence 54.2 ± 7.4 64.1 ± 7.4
(4D)
53.6 ± 7.3
(3D)
Arousal 59.8 ± 7.3 66.1 ± 7.4
(3D)
59.5 ± 7.1
(2D)
It is obvious that the NCA method outperforms
the others. The rest of the experiments are related to
LOSO validation. Table 6 to 8 summarizes these
experiments for valence and arousal.
Table 6: Accuracy after applying LDA DR for valence and
arousal in LOSO validation.
Dimensionality Valence Arousal
2D 56.5 ± 10.7
60.6 ± 9.1
The accuracies for both valence and arousal based
on LOSO validation reveal the same pattern as in 10-
fold cross validation (see Table 1), i.e. accuracy for
arousal is higher than that for valence. These
accuracies are also close to ones in Table 1. For
valence, the result came from the same window size
and overlap parameters in the spectrogram analysis,
but not for arousal.
By comparing Table 2 and Table 7, one can
observe that the best result from arousal came from
the same dimensionality. Looking into detail of the
experiments, one finds out that the best result also
came using the same window size and overlap
parameters in the spectrogram analysis. However, the
valence did not show this pattern.
Table 7: Accuracy after applying NCA DR for valence and
arousal in LOSO validation.
Dimensionality Valence Arousal
2D 61.7 ± 14.1 69.6 ± 12.4
3D 59.4 ± 11.6 51.1 ± 9.5
4D 44.0 ± 12.0 53.3 ± 11.0
5D 40.1 ± 12.0 47.3 ± 11.9
6D 40.0 ± 13.0 51.5 ± 8.6
7D 38.7 ± 11.1 45.7 ± 12.3
Table 8: Accuracy after applying MCML DR for valence
and arousal in LOSO validation.
Dimensionality Valence Arousal
2D 55.9 ± 9.3 61.7 ± 12.3
3D 56.3 ± 12.1 50.2 ± 9.8
4D 41.9 ± 10.6 50.2 ± 10.0
5D 38.8 ± 10.6 50.5 ± 10.4
6D 39.3 ± 11.0 50.3 ± 10.5
7D 39.1 ± 10.8 48.4 ± 8.9
Similar to the NCA result, the accuracy for
arousal also came from the same dimensionality and
parameters of the spectrogram analysis, but not for
valence.
Table 9: Accuracies of all dimensionality reduction
methods in LOSO validation.
LDA NCA MCML
Valence 56.5 ± 10.7 61.7 ± 14.1 56.3 ± 12.1
Arousal 60.6 ± 9.1 69.6 ± 12.4 61.7 ± 12.3
Significance assessment was performed using t-
test with significance level 0.05 for valence between
LDA and NCA methods. The p-value was 0.035
indicating that NCA was superior to LDA. For
arousal, the test showed (p-value 0.0016) that NCA
was superior to MCML.
5 DISCUSSION
As mentioned in the Supervised Dimensionality
Reduction section, DR with the LDA has a limitation
that it can only reduce the dimensionality to a number
not higher than the number of the classes. The other
algorithms can try to search for any dimensionality as
long as it is smaller than the dimensionality of the
original feature space. With this limitation, the LDA
did not provide any improvement for the accuracy but
ICPRAM 2017 - 6th International Conference on Pattern Recognition Applications and Methods
116

can only save some storage space and computational
load.
The MCML, inspired by the NCA (Globerson and
Roweis, 2006), most of the time failed to find the
optimum projection matrix for the features. There
was no improvement to the accuracy of the system
compared to using the original feature set. It reduced
the dimensionality from four to three for valence and
from three to two for arousal. Similar to the LDA, the
contribution of the MCML is saving the storage space
slightly.
The NCA significantly improved the accuracy of
emotion recognition. The dimensionalities of the
feature set were reduced from twenty-three to four
and from twenty-two to three for valence and arousal,
respectively. For small number of samples, this might
be not significant but it will be different for the big
data analysis.
The result of this study is compared to the
accuracies from the previous study (Ferdinando, et
al., 2016), see Table 10.
Table 10: Comparison of results to a reference paper, 10-
fold cross validation.
Reference
(Ferdinando, et al.,
2016)
DR experiment
(NCA)
Valence 55.8 ± 7.3 64.1 ± 7.4 (4D)
Arousal 59.7 ± 7.0 66.1 ± 7.4 (3D)
We verify whether applying DR to features indeed
improves the accuracy of the system using t-test
method with significant level 0.05 and null
hypothesis that both are from the same distribution.
The p-values for both valence and arousal are close to
zero indicating that the improvements are significant.
Table 11: Comparison of results to a reference paper, LOSO
validation.
Original
(Ferdinando, et al.,
2016)
DR experiment
(NCA)
Valence 59.2 ± 11.4 61.7 ± 14.1
Arousal 58.7 ± 9.1 69.6 ± 12.4
We used t-test again to verify that applying DR
can improve the performance of the system in LOSO
validation with significant level 0.05. The p-values
were 0.1873 and 0.0001 for valence and arousal,
respectively, indicating that there is no significant
difference between the original and DR experiment
for valence but there is a significant improvement
with the arousal recognition.
During this study, the algorithms were modified
such that they are iterative with a simple stopping
criterion. Further studies related to iterative
algorithms is needed in order to get more benefits
from the supervised dimensionality reduction. It
might be possible also to investigate how to initialize
matrix
A without random number generator.
6 CONCLUSIONS
This paper explored supervised DR in emotion
recognition based on the Mahnob-HCI database. It
was shown that the supervised DR based on NCA
increased the accuracy from 55.8% to 64.1% and
from 59.7% to 66.1% for a 3-class problem in valence
and arousal respectively using 10-fold cross
validation. Compared to the initial baseline
(Ferdinando, et al., 2014), the accuracies improved
significantly by around 20%.
With LOSO validation, the supervised DR based
on NCA increased the accuracy of arousal recognition
from 58.7% to 69.6% for 3-class problem. However,
it failed to improve the accuracy for valence as
indicated by statistical significance test.
The generalisability of these results is subject to
certain limitations. For instances, the iterative
algorithm was very simple such that the whole system
failed to gain more benefits from the supervised
dimensionality reduction techniques. Another
important limitation is about matrix
A initialization
process which used random number generator. Using
a more sophisticated initialization might improve the
performance.
Among the three methods explored in this paper,
the NCA showed its superiority when it was applied
to the Mahnob-HCI database, although the MCML
was developed to improve the performance of the
NCA. Yet, it will be very interesting to explore the
same methods with other databases and various
applications in order to draw more comprehensive
conclusions for the supervised DR applied to emotion
recognition based on physiological signals.
ACKNOWLEDGEMENTS
This research was supported by by the Directorate
General of Higher Education, Ministry of Higher
Education and Research, Republic of Indonesia, No.
2142/E4.4/K/2013, the Finnish Cultural Foundation
North Ostrobothnia Regional Fund and the
Optoelectronics and Measurement Techniques unit,
University of Oulu, Finland.
Enhancing Emotion Recognition from ECG Signals using Supervised Dimensionality Reduction
117

REFERENCES
Agrafioti, F., Hatzinakos, D. & Anderson, A. K., 2012.
ECG Pattern Analysis for Emotion Detection. IEEE
Transactions on Affective Computing, 3(1), pp. 102-
115.
Ferdinando, H., Seppänen, T. & Alasaarela, E., 2016.
Comparing Features from ECG Pattern and HRV
Analysis for Emotion Recognition System. Chiang Mai,
Thailand, The annual IEEE International Conference on
Computational Intelligence in Bioinformatics and
Computational Biology (CIBCB 2016).
Ferdinando, H., Ye, L., Seppänen, T. & Alasaarela, E.,
2014. Emotion Recognition by Heart Rate Variability.
Australian Journal of Basic and Applied Sciences,
8(14), pp. 50-55.
Fratini, A., Sansone, M., Bifulco, P. & Cesarelli, M., 2015.
Individual identification via electrocardiogram
analysis. BioMedical Engineering OnLine, 14(78), pp.
1-23.
Globerson, A. & Roweis, S., 2006. Metric Learning by
Collapsing Classes. In: Y. Weiss & B. Schölkopf, eds.
Advances in Neural Information Processing Systems
18. Cambridge, MA: MIT Press, p. 451–458.
Goldberger, J., Roweis, S., Hinton, G. & Salakhutdinov, R.,
2005. Neighborhood Components Analysis. In: L. K.
Saul, Y. Weiss & L. Bottou, eds. Advances in Neural
Information Processing System Vol. 17. Cambridge:
MIT Press, p. 513–520.
Jolliffe, I., 2002. Principal Component Analysis. 2 ed. New
York: Springer Verlag.
Labiak, J. & Livescu, K., 2011. Nearest Neighbors with
Learned Distances for Phonetic Frame Classification.
Florence, Italy., International Speech Communication
Association (ISCA).
Lee, J. A. & Verleysen, M., 2010. Unsupervised
Dimensionality Reduction: Overview and Recent
Advances. Barcelona, Spain, IEEE World Congress on
Computational Intelligence (WCCI) 2010.
McDuff, D. et al., 2012. AffectAura: an intelligent system
for emotional memory. New York, Association for
Computing Machinery (ACM).
McSharry, P. E., Clifford, G. D., Tarassenko, L. & Smith,
L. A., 2003. A Dynamical Model of Generating
Synthetic Electrocardiogram Signals. IEEE
Transactions on Biomedical Engineering, 50(3), pp.
289-294.
Rilling, G., Flandrin, P., Gonçalves, P. & Lilly, J. M., 2007.
Bivariate Empirical Mode Decomposition. IEEE Signal
Processing Letters, 14(12), pp. 936-939.
Romero, J., Diago, L., Shinoda, J. & Hagiwara, I., 2015.
Comparison of Data Reduction Methods for the
Analysis of Iyashi Expressions using Brain Signals.
Journal of Advanced Simulation in Science and
Engineering, 2(2), pp. 349-366.
Sammon, J. W., 1969. A nonlinear mapping algorithm for
data structure analysis. EEE Transactions on
Computers, CC-18(5), pp. 401-409.
Soleymani, M., Lichtenauer, J., Pun, T. & Pantic, M., 2012.
A Multimodal Database for Affect Recognition and
Implicit Tagging. IEEE Transactions on Affective
Computing, 3(1), pp. 1-14.
Sugiyama, M., 2007. Dimensionality Reduction of
Multimodal Labeled Data by Local Fisher Discriminant
Analysis. Journal of Machine Learning Research,
Volume 8, pp. 1027-1061.
Valenzi, S., Islam, T., Jurica, P. & Cichocki, A., 2014.
Individual Classification of Emotions Using EEG.
Journal of Biomedical Science and Engineering,
Volume 7, pp. 604-620.
van der Maaten, L., 2016. Matlab Toolbox for
Dimensionality Reduction - Laurens van der Maaten.
[Online]
Available at: https://lvdmaaten.github.io/drtoolbox/
[Accessed 28 7 2016].
Weinberger, K. Q., Blitzer, J. & Saul, L. K., 2005. Distance
Metric Learning for Large Margin Nearest Neighbor
Classification. Advances in Neural Information
Processing System, Volume 18, p. 1473–1480.
Weinberger, K. Q. & Saul, L. K., 2009. Distance Metric
Learning for Large Margin Nearest Neighbor
Classification. Journal of Machine Learning Research,
Volume 10, pp. 207-244.
Zhang, S. & Zhao, X., 2013. Dimensionality reduction-
based spoken emotion recognition. Multimedia Tools
and Applications, 63(3), p. 615–646.
ICPRAM 2017 - 6th International Conference on Pattern Recognition Applications and Methods
118
