
DNA Analysis: Principles and Sequencing Algorithms
Veronika Abramova
1
, Bruno Cabral
2
and Jorge Bernardino
1,2
1
Polytechnic Institute of Coimbra, ISEC - Coimbra Institute of Engineering, Coimbra, Portugal
2
University of Coimbra – CISUC, Centre for Informatics and Systems of University Coimbra, Coimbra, Portugal
Keywords: DNA, Genome Assembly, Graph, Assemblers, Algorithms.
Abstract: DNA discovery has put humans one step closer to deciphering their own structure stored as biological data.
Such data could provide us with a huge amount of information, necessary for studying ourselves and learn
all the variants that pre-determine one's characteristics. Although, these days, we are able to extract DNA
from our cells and transform it into sequences, there is still a long road ahead since DNA has not been easy
to process or even extract in one go. Over the past years, bioinformatics has been evolving more and more,
constantly aiding biologists on the attempts to “break” the code. In this paper, we present some of the most
relevant algorithms and principles applied on the analysis of our DNA. We attempt to provide basic genome
overview but, moreover, the focus of our study is on assembly, one of the main phases of DNA analysis.
1 INTRODUCTION
Deoxyribonucleic acid, known as DNA, was firstly
discovered and introduced by Watson and Crick
(Watson and Crick, 1953). Authors stated that any
existing organism’s molecule stores all the necessary
data for its living and grown by carrying instructions
that are necessary for constant development and
overall organism’s functioning (Saenger and
Wolfram, 1984), (Alberts et. al, 2002). For us,
humans, DNA contains most of our characteristics
as a person, such as, adaptation, character and so on
but, more importantly, information about creation of
proteins that form all of our parts. This protein is
used by our organism to produce all the necessary
substances, for example, keratin or our hair and
nails, bio functioning proteins that transport oxygen
inside our body and so on. DNA structure is
presented as double helix, where on one helix are
located Adenine (A) and Guanine (G) and on the
other helix Thymine (T) and Cytosine (C). While
creating the double helix, A connects with T and,
respectively, C with G. Those are called base pairs.
Currently, there are several challenges related to
the DNA study and manipulation. One of the
limitations of the current technology is the capability
of extracting the entire DNA chain, only parts of
DNA may be read. That means that, these days, it is
not possible to extract the entire genome. More than
that, extracted parts can be easily corrupted, either
by extraction errors or by transcription errors.
Moreover, even if the part of DNA that is analyzed
was correctly processed, there is a possibility of that
sequence being just a part of necessary sequence or
not meaning anything (trash). That means that even
if the code was extracted correctly, the extracted
sequence may not have expected importance.
Another challenging task that follows is genome
assembly.
In this paper we attempt describing some of the
challenges that are related to the genome assembly
as well as some of the current possibilities. Currently
there are a lot of studies and papers, focusing on
extracting ever more of the necessary and important
data. However, we consider that it may be
challenging for someone to begin study
bioinformatics. Therefore, we describe the
assembling algorithms and the origin of those as
well as their base, so readers can understand some of
the associated challenges with genome sequencing.
The remaining of the paper is structured as
follows: Section 2 describes related work and some
of the studies that have been performed over the past
years. Section 3 describes some of the available
assemblers that are used for extracting and
sequencing DNA. Section 4 provides an overview
over algorithms that are used for DNA sequencing
(genome assembly). Finally, section 5 presents our
conclusions and future work.
Abramova, V., Cabral, B. and Bernardino, J.
DNA Analysis: Principles and Sequencing Algorithms.
DOI: 10.5220/0006084102450250
In Proceedings of the 8th International Joint Conference on Computational Intelligence (IJCCI 2016) - Volume 1: ECTA, pages 245-250
ISBN: 978-989-758-201-1
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
245

2 RELATED WORK
DNA study has been a hardly invested area over the
past years and, therefore, there is a lot of data about
genetic study and evolution. Munib et al. (2015)
present and describe the entire process of shotgun
sequence assembly in the realm of high-performance
computing. They describe main phases of genome
assemble and some of the algorithms that may be
used among those phases. Also, Mount (2004)
provides an examination of the computational
methods needed for analyzing DNA, RNA –
structure that DNA is transformed after has been
extracted from the nucleus of the cell, and protein
data, as well as genomes. Differently, we focus our
paper on one specific area, providing more detail,
instead of entire process overview. Polyanovsky et
al. (2011) present a comparison approach between
alignment algorithms. Authors based they work
mainly on Polyanovsky et al. (2008), Sunyaev et al.
(2004), Domingues et al. (2000) and compared
results produced by local and global alignment and
concluded that both local and the global alignment
algorithms rely on positions and relative lengths of
the parts of the sequences that will be aligned and
depending of the sequence positioning, global or
local algorithms provide better results. Global
algorithm proved to that when the core part of one
sequence was positioned above the core of the other
sequence but when the cores were positioned
asymmetrically, the local algorithm was more stable.
Posada (2009) describes methods to analyze DNA
sequences as well as some available tools that are
free to use, describing recognition of similar
sequences using BLAST, sequence alignment, DNA
compositions and molecular evolution and other.
This book is more oriented for biologists that do not
have much knowledge about computer science and,
moreover, have a bigger biological background that
most of computer engineers do not have. Jones and
Pavel (2004
) provide a large overview over existing
algorithms that are used in bioinformatics by
presenting the foundations of algorithms, describing
principles that drove their design, and their most
important results. DNA sequencing challenges and
processing are described in El-Metwally et al.
(2013), Niedringhaus et al. (2011), Voelkerding et
al. (2009), Zhou et al. (2010), Wheeler et al. (2008).
Authors describe some of the past and current
technologies, solutions for DNA sequencing and
available next-generation sequencing (NGS)
platforms and their future (Hert et al., 2008). NGS
platforms perform sequencing of millions of small
fragments of DNA in parallel which means that
millions of small fragments of DNA can be
sequenced at the same time, creating a massive pool
of data. All the sequence fragments are analysed
several times in order to reduce error possibility.
3 GENOME ASSEMBLERS
After years of genome study and research there is a
variety of assemblers used to extract and transcript
genome. Constant interest and technological
evolution have been contributing for different
generations of assemblers, attempting to provide
better results and surpass each other. The first next-
generation DNA sequencing machine, the GS20,
was introduced to the market by 454 Life Sciences
in 2005 (Gilles et al., 2011) That technology was
based on a large-scale parallel pyrosequencing
system, which relies on fixing DNA fragments to
small DNA-capture beads in a water-in-oil emulsion.
While trying to understand what is genome
assembling and what its stages are and elements that
are take part, one of the first things we hear of is de-
novo assembly. So what is de-novo how it is
important? De-novo is a method of new generation
sequencing that attempts to sequence genome
without any reference. That means that short reads
are not mapped against original sequence (when it is
available). On the other hand, reference assembling
considers available parts of the sequence (reads) and
only considers those that can be mapped into an
original sequence. It may be used to assemble
genomes of already extinct organisms (Era7, 2016).
One of the first challenges consisted of the
extractions of the DNA from the cells. Biological
advance and evolution had contributed to this task
and currently it is possible to extract DNA. That
possibility would provide all of the answers to one’s
“construction” and allow us to manipulate our own
constituent and fight many diseases. DNA structure
is presented as double helix, where on one helix are
located Adenine (A) and Guanine (G) and on the
other helix Thymine (T) and Cytosine (C). While
creating the double helix, A connects with T and,
respectively, C with G. Those are called base pairs.
After extracting a DNA sequence (input), it is
processed by overlapping the extracted sequences.
This overlap process is called sequence alignment
and it allows creating a larger sequence that contains
parts of small pieces. The output of an assembler is
generally decomposed into contigs, or contiguous
regions of the genome which are nearly completely
resolved, and scaffolds, or sets of contigs which are
approximately placed and oriented with respect to
ECTA 2016 - 8th International Conference on Evolutionary Computation Theory and Applications
246

each other (Gregory, 2005). Currently DNA
sequencing technology allows short reads of 500 and
up to 700 nucleotides. This sequences are parts from
one complete DNA genome.
Genome assembling from shorter sequences is
like reassembling the magazine from the millions of
small pieces of each page. In both cases will exist
errors. In case of magazine it would be ragged edges
that difficult putting pieces together. In case of
genome assembly those would be reading errors.
More than that, pieces may be lost in both cases and
also, in case of DNA, nucleotides may be
transcripted incorrectly. Nevertheless, efforts to
determine the DNA sequence of organisms have
been remarkably successful, even in the face of these
difficulties. Salzberg et al. (2011) describe some of
the most popular Assemblers.
4 ALGORITHMS
In the 1950s, Seymour Benzer applied graph theory
to show that genes are linear (Jones and Pevzner,
2004). Therefore, in this section we will describe
some of the most popular algorithms for genome
assembly, while most of those being Graph
algorithms. Short read assemblers generally use one
of two basic algorithms: overlap graphs and de
Bruijn graphs. The overlaps between each pair of
reads is computed and compiled into a graph, in
which each node represents a single sequence read.
This algorithm is more computationally intensive
than de Bruijn graphs, and most effective in
assembling fewer reads with a high degree of
overlap (Illumina¸ 2010). De Brujin Graph started
from Bridges of Königsberg problem (Compeau et
al., 2011). The bridges problem is presented on the
Figure 1.
Figure 1: Bridges of Königsberg (Compeau et al.,2011).
The problem statement consisted on passing
each bridge only once considering that the islands
could only be reached by the bridges and every
bridge once accessed must be crossed to its other
end. The starting and ending points of the route may
or may not be the same. This problem was firstly
solved by Leonard Euler, a Swiss mathematician,
physicist, astronomer and logician. At first Euler
analysed the problem and decided that it could not
be solved (Paoletti, 2011). However, Euler could
not just accept that. He decided to create a logical
formula for this type of problems. Euler observed
that in order to be able to solve this type of problems
it would be necessary zero or two nodes with odd
degree. This theory was posteriorly proved by Carl
Hierholzer. This author stated that in order to be able
to solve a graph using Euler’s path, each connected
node has to have an even degree, except for the
starting and terminal nodes (Barnett, 2009). On
August 26, 1735, Euler presented a paper containing
the solution to the Konigsberg bridge problem. He
addressed Bridges of Königsberg, problem as well
as provided a general solution where any number of
bridges could be used (Hopkins and Wilson, 2004).
In this section we describe some of the basic
DNA assembly algorithms and problems that those
try to solve.
4.1 Shortest Superstring Problem
Since all the DNA reads are performed as parts, it is
necessary to be able to overlap those and create one
superstring that would better represent and include
considered pieces of sequences. This is important
since different sequences may be part of the same
superstring. However, this superstring creation may
be challenging. As more simplistic approach,
superstring could result from complete
concatenation of available reads. But this approach
would not be very correct. Best superstring created
should be the shortest one possible. That would
represent the sequence that somehow concatenates
available reads but, also, does not contain
unnecessary information.
INPUTS: Set of sequences.
OUTPUT: One sequence that contains all the
input sequences while being as short as possible.
In order to be able to create one superstring it is
necessary to align input sequences and be able to
identify which ones may be same part of different
reads. We should be able to define any overlapping
regions and after that create one complete sequence.
Below we present an example of Shortest
Superstring Solution. Given:
INPUTS: {00, 01, 10, 11}
FULL SEQUENCE: 00011011
SHORTEST SUPERSTRING:
00
DNA Analysis: Principles and Sequencing Algorithms
247
01
10
11
001011
As it is possible to understand, instead of just
concatenating all the available stings, in order to
solve Shortest Superstring Problem, it is necessary
to evaluate all the sequences and find overlaps. After
those overlaps are considered, final sequence is
shorter than just full sequence concatenation.
As computational approach, in order to solve
these problems may be used greedy algorithms
(Vazirani¸ 2001). Those algorithms continuously
build up the final solution, through interactive
cycles. Each cycle the algorithm attempts to find
optimal solution. It is important to notice that greedy
algorithms are not exhaustive and when the problem
has considerable difficulty it may not be possible to
obtain the best optimal solution, instead just local
maximum. Also, depending on the problem
difficulty, those algorithms require a considerable
amount of cycles in order to be able to produce
optimal solution that may, never less, be local.
While applied to the Shortest Superstring Problem,
greedy algorithms iterate input sequences and
integrate (assemble) those one by one. In the
approach presented below the algorithm considers
two of the most similar sequences and overlap them,
creating just one sequence. After, this sequence will
be aligned with the third one and so on, for example:
INPUT: {ACTG, CTGA, GACC, TGAC,
ACCC}
STAGE 1 – consider most similar string ACTG
and CTGA
ACTG
CTGA -> ACTGA
STAGE 2 – consider most similar ACTGA and
TGAC
ACTGA
TGAC -> ACTGAC
STAGE 3 – consider most similar ACTGAC
and GACC
ACTGAC
GACC -> ACTGACC
STAGE 4 – overlap the rest – ACTGACC and
ACCC
ACTGACC
ACCC -> ACTGACCC
OUTPUT: ACTGACCC
As we can see, in the previous example, greedy
algorithm was able to create a shortest superstring
with 4 steps. However, the problem presented is
very simplistic. In real case scenario, inputs string
would have been larger and number of inputs would
be bigger. Therefore, it may be difficult for the
algorithm to find the best superstring. Also, the
algorithm may consider different possibilities of
sequences to match, especially at start. This also
highly affects the final sequence.
4.2 Hamiltonian Path Problem
Hamiltonian Path in an undirected graph is a path
that visits each vertex exactly once. A cycle that
uses every vertex in a graph exactly once is called a
Hamilton cycle. It is important to notice that in order
for Hamiltonian Path to exist, graph edges must be
biconnected. That means that two or more vertices
of the graph when removed could not disconnect the
graph. Problem statement is, as follows:
INPUT: Read sequences
OUTPUT: Created graph as well as Spectrum S
Available sequences must be connected if there is an
overlap. For example,
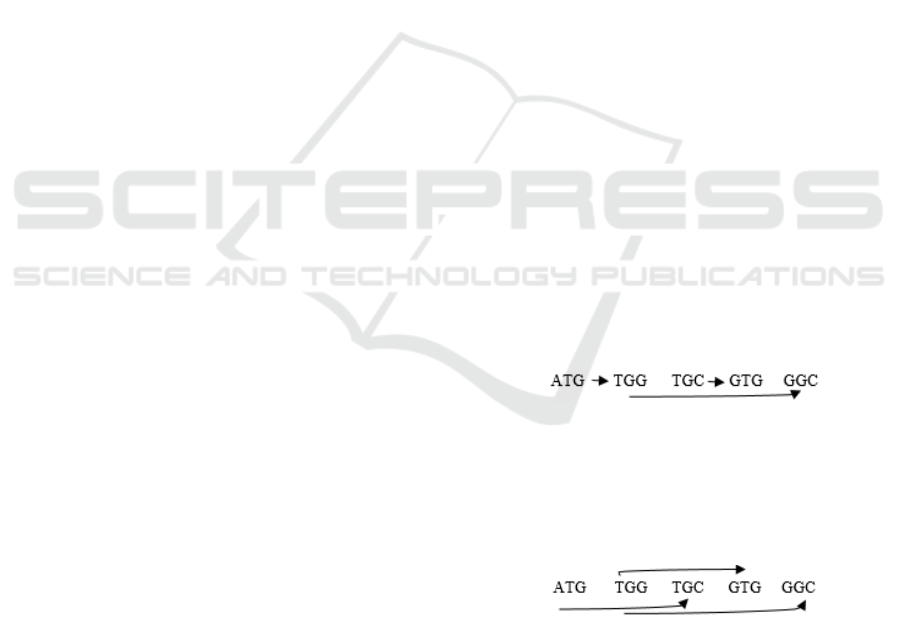
INPUT: {ATG, TGG, TGC, GTG, GGC}
OUTPUT:
ATGGCGTGC
In this example there is a possibility of the
second path that could be obtained by overlapping
sequences in different order (way):
OUTPUT:
ATGCGTGGC
This simple example allows us to create different
path by overlapping existing sequences. Due to the
nature of the sequences (available nucleotides) there
were two possible path to choose from. However, in
the more complex problem the solution is not a
trivial one.
ECTA 2016 - 8th International Conference on Evolutionary Computation Theory and Applications
248

As first approach could be used brute-force
(exhaustive) search. But, a better approach is
presented by Frank Rubin Author proposed an
algorithm that divides a complete problem into
pieces, each being solved easier than them all
together (Rubin, 1974). All the available graphs are
divided into three categories: used in path, not used
in path and undecided. The functioning of this
algorithm (identification and study of small
problems) resembles dynamic programming
principles, where each stage of the problem (part) is
independent from previous stages (Vazirani¸2001).
Also, Michael Held and Richard M. Karp
presented their solution using dynamic programming
and based on the traveling-salesman problem (Held
and Karp, 1962). Traveling-salesman problem is a
well knows problem, similarly to the Bridges of
Königsberg problem. Salesman must visit each city
just once and using shortest route. Therefore, authors
provided a solution as cyclic iteration of vertices and
verify if there is a possibility of covering a path in
set of vertices S. For further vertices the path is
evaluated only if it existed in the previous stage.
4.3 Eulerian Path Problem
Similarly, to the previously stated problem, the
purpose of the Eulerian Path is to find a route to visit
each node of the graph just once. Euler Circuit is a
circuit that uses each vertex of the graph just once.
The main difference between Eulerian path and
Circuit is that path ends on the different vertex,
while circuit represents the complete cycle where it
starts and end with the same vertex. Differently from
the Hamiltonian Path problem, while working with
Eulerian Path, we shall consider more linear
traveling approach. That means that sequences are
connected linearly (v1 -> v2 -> …) which not
happens in Hamiltonian Path. Moreover, while
working with Eulerian path it is necessary to
consider the following theorem: “A connected graph
is Eulerian if and only if each of its vertices is
balanced.” (Jones and Pevzner
, 2004). Consider that
vertex is balanced when indegree = outdegree for
presented vertex. In order to create a Eulerian cycle,
it is necessary to start drawing path from the
arbitrary vertex v and traversing edges that have not
already been used. We stop the path when we
encounter a vertex with no way out, that is, a vertex
whose outgoing edges has already been used in the
path.
Below is presented an example of the Eulerian
Path problem:
INPUT: {ATG, TGG, TGC, GTG, GGC}
STAGE 1: Create vertices of lengh k-1
{AT, TG, GG, GC, CG, GT}
OUTPUT:
ATGGCGTGC
As we have seen in the previous sub-section,
there is another path:
OUTPUT:
ATGCGTGGC
In order to solve Eulerian path problems there are
two main algorithms: Fleury's algorithm and Cycle
finding algorithm. Fleury's algorithm was originally
created in 1883 to find Eulerian paths (Fleury,
1883). It proceeds by repeatedly removing edges
from the graph in such way, that the graph remains
Eulerian. The algorithm starts at a vertex of odd
degree, or, if the graph has none, it starts with an
arbitrarily chosen vertex. At each step it chooses the
next edge in the path to be removed considering that
that edge would not disconnect the graph itself (is
not a bridge). If there is no such edge algorithm
cannot proceed. It then moves to the other endpoint
of that vertex and deletes the chosen edge. At the
end of the algorithm there are no edges left, and the
sequence from which the edges were chosen
(removed) forms a Eulerian cycle if the graph has no
vertices of odd degree, or a Eulerian trail if there are
exactly two vertices of odd degree.
5 CONCLUSIONS
This work provides a basic overview over genome
assembly processes and their challenges. We started
by describing the overall genome sequencing
problem and discussing its relevance. Posteriorly,
we portrayed some of the most popular assemblers
and their inner workings. Most of the assemblers,
including some not mentioned in this work, use de
Bruijn graphs for sequence assembling. But,
DNA Analysis: Principles and Sequencing Algorithms
249

sequence alignment, used for creating common
substrings (overlaps) also has an high impact and
weight in DNA sequencing performance. Finally, to
provide a clear insight for computer scientists into
genome assembly algorithms, we draw some
parallelism between the Selling-travelers problem,
as well as with the Bridges of Königsberg problem,
with the genome sequencing problems, showing that
both have highly contributed to the evolution and the
development of graph algorithms and their use in
bioinformatics.
After many years of research and work, genome
assembly is still a hard and cumbersome task. As
future work, we plan to implement and optimize
some of the described approaches using state of the
art parallel and distributed computing strategies.
REFERENCES
Alberts, B., Johnson, A., Lewis,J., Raff, M., Roberts, K.,
Walters, P. 2002. Molecular Biology of the Cell;
Fourth Edition. New York and London: Garland
Science. ISBN 0-8153-3218-1.
Barnett, J. H. 2009. Early Writings on Graph Theory:
Euler Circuits and The Königsberg Bridge Problem.
Compeau, P. E. C., Pevzner, P. A. and Tesler, G. 2011.
How to apply de Bruijn graphs to genome assembly.
Domingues, F.S., Lackner, P., Andreeva, A., Sippl, M.J.
2000. Structure-based evaluation of squence coparison
and fold recognition alignment accuracy. J Mol Biol.
2000;297:1003–1013.
El-Metwally, S., Taher, H., Magdi, Z. and Helmy, M.
2013. Next-Generation Sequence Assembly: Four
Stages of Data Processing and Computational
Challenges. PLoS Comput Biol. 2013 Dec; 9(12):
e1003345.
Era7. 2016. https://era7bioinformatics.com/en/page.cfm?
id=1500 retrieved on 20.01.2016.
Fleury, M. 1883. Deux problèmes de Géométrie de
situation. Journal de mathématiques élémentaires, 2nd
ser. (in French) 2: 257–261.
Gilles, A., Meglécz, E., Pech, N., Ferreira, S., Malausa, T.,
Martin, J.F. Accuracy and quality assessment of 454
GS-FLX Titanium pyrosequencing, 2011, 12:245.
Gregory, S. 2005. Contig Assembly. Encyclopedia of Life
Sciences.
Held, M. and Karp, R. M. 1962. A dynamic programming
approach to sequencing problems. J. Siam 10 (1):
196–210.
Hert, D.G., Fredlake, C.P., Barron, A.E. 2008. Advantages
and limitations of next-generation sequencing
technologies: a comparison of electrophoresis and
non-electrophoresis methods. Electrophoresis. 29(23):
4618-26.
Hopkins, B, and Wilson, R. The Truth about Königsberg.
College Mathematics Journal (2004), 35, 198-207.
Illumina, Inc. 2010. De Novo Assembly Using Illumina
Reads. Nature 171:737–738.
Mount, DM. 2004. Bioinformatics: Sequence and Genome
Analysis (2nd ed.). Cold Spring Harbor Laboratory
Press: Cold Spring Harbor, NY. ISBN 0-87969-608-7.
Munib, A., Ishfaq, A., Mohammad S. A. 2015. A survey
of genome sequence assembly techniques and
algorithms using high-performance computing. The
Journal of Supercomputing. Vol. 71 (1), pp 293-339.
Jones, N.C. and Pevzner, P.A. 2004. An Introduction to
Bioinformatics Algorithms. © 2004 Massachusetts
Institute of Technology.
Niedringhaus, T.P., Milanova, D., Kerby, M.B., Snyder,
M.P., Barron, A.E. 2011 Landscape of next-generation
sequencing technologies. Anal Chem 83: 4327– 4341.
Paoletti, T. 2011. Leonard Euler's Solution to the
Konigsberg Bridge Problem. Convergence 2011.
Polyanovsky, V. O., Roytberg, M. A., Tumanyan, V. G.
2011. Comparative analysis of the quality of a global
algorithm and a local algorithm for alignment of two
sequences. Algorithms for Molecular Biology 6.
Polyanovsky, V., Roytberg, M.A., Tumanyan, V.G. 2008.
Reconstruction of genuine pair-wise sequence
alignment. J Comp Biol. 2008;15:379–391.
Posada, D. 2009. Bioinformatics for DNA Sequence
Analysis. (Ed.). ISBN 978-1-59745-251-9.
Rubin, F. 1974. A Search Procedure for Hamilton Paths
and Circuits. Journal of the ACM 21 (4): 576–80.
Saenger, W. 1984. Principles of Nucleic Acid Structure.
New York: Springer-Verlag. ISBN 0-387-90762-9.
Salzberg, S. L., Phillippy, A.M., Zimin, A., Puiu1, D.,
Magoc, T., Koren, S., Treangen, TJ., Schatz, M.C.,
Delcher, A.L., Roberts, M., Marçais, G., Pop, M. and
Yorke, J.A. 2011. GAGE: A critical evaluation of
genome assemblies and assembly algorithms.
Sunyaev, S.R., Bogopolsky, G.A., Oleynikova, N.V.,
Vlasov, P.K., Finkelstein, A.V., Roytberg, M.A. 2004.
From analysis of protein structural alignments toward
a novel approach to align protein sequences. Proteins:
Structure, Function and Bioinforrmatics. 2004;
54:569–582.
Vazirani, U. V. 2001. Algorithms.
Voelkerding, K.V., Dames, S.A., Durtschi, J.D. 2009.
Next-generation sequencing: from basic research to
diagnostics. Clin Chem 55: 641–658.
Watson J, Crick. 1953. Molecular structure of nucleic
acids: a structure for deoxyribose nucleic acid.
Wheeler, DA, Srinivasan, M., Egholm, M., Shen, Y.,
Chen, L., McGuire, A. 2008. The complete genome of
an individual by massively parallel DNA sequencing.
Nature 2008; 452:872-876.
Zhou, X., Ren, L., Meng, Q., Li, Y., Yu, Y. 2010. The
next-generation sequencing technology and
application. Protein Cell 1: 520–536.
ECTA 2016 - 8th International Conference on Evolutionary Computation Theory and Applications
250
