
Combining Bayesian Approaches and Evolutionary Techniques for the
Inference of Breast Cancer Networks
Stefano Beretta
1
, Mauro Castelli
2
, Ivo Gonc¸alves
2
, Ivan Merelli
3
and Daniele Ramazzotti
4
1
DISCo, Universit
´
a degli Studi di Milano Bicocca, 20126 Milano, Italy
2
NOVA IMS, Universidade Nova de Lisboa, 1070-312 Lisboa, Portugal
3
Ist. di Tecnologie Biomediche, Consiglio Nazionale delle Ricerche, Segrate, Italy
4
Department of Pathology, Stanford University, Stanford, U.S.A.
Keywords:
Bayesian Graphical Models, Breast Cancer, Genetic Algorithms, Network Inference.
Abstract:
Gene and protein networks are very important to model complex large-scale systems in molecular biology.
Inferring or reverseengineering such networks can be defined as the process of identifying gene/protein inter-
actions from experimental data through computational analysis. However, this task is typically complicated
by the enormously large scale of the unknowns in a rather small sample size. Furthermore, when the goal is
to study causal relationships within the network, tools capable of overcoming the limitations of correlation
networks are required. In this work, we make use of Bayesian Graphical Models to attach this problem and,
specifically, we perform a comparative study of different state-of-the-art heuristics, analyzing their perfor-
mance in inferring the structure of the Bayesian Network from breast cancer data.
1 INTRODUCTION
Molecular networks are essential for every biologi-
cal process, since genes and proteins are able to carry
out their function only in precisely regulated path-
ways. For this reason, data-driven learning of regula-
tory connections in molecular networks has long been
a key topic in computational biology (Bansal et al.,
2007). The general problem is to infer, or reverse-
engineer, from gene or protein expression data, the
regulatory interactions among these biological enti-
ties using computational algorithms.
In this context, despite correlation networks are
widely used for gene expression and proteomic data
analysis, it is known that correlations not only con-
found direct and indirect associations, but also pro-
vide no means to distinguish between cause and ef-
fect. For causal analysis the inference of a directed
graphical model is typically required. However, this
task is rather difficult due to multiple theoretical and
practical reasons, among which, but not limited to, the
course of dimensionality (Pearl, 2003).
Therefore, causal analysis requires tools capable
of overcoming the limitations of correlation networks:
much of the work in this area has focused on Bayesian
Networks (Pearl, 2003) or related regression models,
such as systems of recursive equations or influence di-
agrams. All these models describe causal relations by
an underlying directed acyclic graph (DAG). Never-
theless, it remains unclear whether causal, rather than
merely correlational, relationships in molecular net-
works can be inferred in complex biological settings.
Moreover, the problem is typically complicated by
the enormously large scale of the unknowns in a rather
small sample size. Furthermore, data is prone to ex-
perimental defects and noisy readings, while many
other biases can compromise the quality of the results.
These complexities call for a heavy involvement of
powerful mathematical models which play an in-
creasingly important role in this research area (Kabir
et al., 2010). In order to assess the ability of dif-
ferent tools to learn causal networks, the Dialogue
for Reverse Engineering Assessment and Methods
(DREAM) project has run several challenges focused
on network inferences (Stolovitzky et al., 2007). In
particular, we focused on (sub)-challenge 8.1 con-
cerning Human Protein Networks (HPN) in cancer
cell lines, which is about the inference of causal sig-
nalling pathways using time-course data with pertur-
bations on network nodes. This sub-challenge was
split into two independent parts, concerning Breast
Cancer proteomic data and in silico data.
Beretta, S., Castelli, M., Gonçalves, I., Merelli, I. and Ramazzotti, D.
Combining Bayesian Approaches and Evolutionary Techniques for the Inference of Breast Cancer Networks.
DOI: 10.5220/0006064102170224
In Proceedings of the 8th International Joint Conference on Computational Intelligence (IJCCI 2016) - Volume 1: ECTA, pages 217-224
ISBN: 978-989-758-201-1
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
217

Different types of models, such as directed graphs,
Boolean networks (Akutsu et al., 1999), Bayesian
Graphical Models (Zou and Conzen, 2005), and var-
ious differential models have been used to describe
gene regulations at various levels of detail and com-
plexity. The choice of the model is often determined
by how much information it tries to capture, taking
into account that the more information a model at-
tempts to infer, the more parameters are needed to
learn it, and the more complex the overall approach
becomes. Specifically, researchers have paid great at-
tention to Bayesian Networks, which can compactly
model dependency relationships between variables
relying on probabilistic measures. Since gene expres-
sion experiments are subject to many measurement er-
rors, the use of statistical methods is expected to be
effective for extracting useful information from such
noisy data. Friedman et al. (Friedman et al., 2000)
proposed both discrete and continuous Bayesian net-
work models relying on linear regression for infer-
ring gene networks. Imoto et al. (Imoto et al., 2001)
succeeded in employing non-parametric regressions
for capturing even non-linear relationships between
genes.
In this work, we perform a comparative study of
different heuristics at the state-of-the-art to perform
the task of inferring the structure of a Bayesian net-
work from breast cancer data. The paper is struc-
tured as follows: Section 2 provides a background of
the biological problem under exam; Section 3 gives
a formal definition of the problem addressed in this
study, along with a description of the different compu-
tational and statistical machineries that we are adopt-
ing, and of the input data. Afterwards, the results of
the described methods on real and simulated data are
presented and discussed in Section 4. Section 5 con-
cludes the paper and suggests avenues for future re-
search.
2 BIOLOGICAL BACKGROUND
Many biological processes are carried out by inter-
actions between proteins, RNA, and DNA. Cells re-
spond to their environment by activating signalling
networks that trigger processes such as growth, sur-
vival, apoptosis (programmed cell death), and migra-
tion. Post-translational modifications, notably phos-
phorylation, play a key role in these signalling events.
In cancer cells, signalling networks frequently be-
come compromised, leading to abnormal behaviours
and responses to external stimuli. Endogenous sig-
nal transduction in cancer cells is systematically dis-
turbed to redirect the cellular decisions from differen-
tiation and apoptosis to proliferation and, later, inva-
sion. Cancer cells acquire their malignancy through
accumulation of advantageous gene mutations by
which the necessary steps to malignancy are obtained.
These selfish adaptations to independence can be de-
scribed as a result from an evolutionary process of di-
versity and selection (Schramm et al., 2010).
Many current and emerging cancer treatments
are designed to block nodes in signalling networks,
thereby altering signalling cascades. Although there
is a wealth of literature describing canonical cell sig-
nalling networks, little is known about exactly how
these networks operate in different cancer cells. Ad-
vancing our understanding of how these networks are
deregulated across cancer cells will ultimately lead to
more effective treatment strategies for patients.
Recently, high-throughput analysis enabled the
possibility to obtain genome-wide information, such
as mRNA expressions, protein-protein interactions,
protein localizations and so on. A lot of attention has
been dedicated on developing computational methods
for extracting valuable information of molecular net-
works from such various types of genomic data.
Currently, statistical models for estimating gene
regulatory networks from genomic data are mainly
based on expression data from DNA microarrays or
RNA-seq experiments. However, since information
from these approaches is limited by their quality,
noise and experimental errors, sophisticated mathe-
matical approaches are necessary for estimating gene
regulatory networks accurately.
On the other hand, protein-protein interaction net-
works are mainly constructed relying on observed
protein-protein interaction data, using approaches
such the two hybrid assays, tandem affinity purifica-
tion experiments and, more recently, protein arrays.
However, protein-protein interaction data often con-
tains some errors, making even more difficult to con-
struct comprehensive protein-protein interaction net-
works from these interaction data alone.
3 METHODS
A Bayesian Network (BN) is a statistical graphical
model that represents a joint distribution over n ran-
dom variables and encodes it by means of a direct
acyclic graph (DAG) depicting the n nodes referring
to the variables. More formally, we define a BN as a
direct acyclic graph G = (V,E), where V is the set
containing the n random variables and E is the set
of the directed arcs over them, representing any con-
ditional dependence among the variables (Parsons,
2011).
ECTA 2016 - 8th International Conference on Evolutionary Computation Theory and Applications
218

In this work, we make use of such graphical tool
to model a protein network G
p
(being a direct acyclic
graph), whose structure (i.e., the nodes and arcs in
the model) maximizes the likelihood, given the ob-
served data on which we make the inference. More-
over, we define this task as an optimization problem
where, for a set of observations D, we aim at max-
imizing the likelihood of observing the data given a
specific model G
p
, which we define as
L L (G
p
,D) =
∏
d∈D
P(d|G
p
),
that is the product of the conditional probabilities
given each observation d ∈ D.
Practically, however, there is a well-known issue
when learning the network structure by maximizing
the likelihood function. In fact, for any arbitrary set of
data, the most likely graph is usually very connected,
since adding an edge typically can only increase the
likelihood of the data, hence leading to overfitting. To
try to reduce this problem, the likelihood is almost
always adjusted by means of a regularization term
that penalizes the complexity of the model (Parsons,
2011).
We also observe that, regardless of the adopted ap-
proach and likelihood score, the main issue to infer
the structure of a BN is the huge search space of the
valid solutions, which makes this a well known NP-
hard problem and, therefore, one will need to make
use of heuristics to perform such inference (Parsons,
2011).
In this work, we compare different heuristics
search algorithms along with various regularizations
for the likelihood score. In Table 1 we present a list
of combinations of the adopted techniques, which are
described in details in the subsequent sections.
Table 1: Combinations of the different heuristics and regu-
larization approaches used in this work.
Heuristic Search Algorithm Regularizators
Hill Climbing (HC) loglik AIC BIC
Tabu Search (TB) loglik AIC BIC
Genetic Algoritms (GA) loglik AIC BIC
Here we employ three different and well-known
evolutionary methods to solve the previously men-
tioned optimization problem, that is to reconstruct the
Bayesian network w.r.t. to a specific regularization
score. In the rest of this section we briefly describe
each method and also the considered regularizators.
3.1 Hill Climbing
Hill Climbing (HC) is one of the simplest iterative
techniques that have been proposed for solving op-
timization problems. While HC consists of a simple
and intuitive sequence of steps, it is a good search
technique to be used as a baseline for comparing
the performance of more advanced optimization tech-
niques.
Hill climbing shares with other techniques
(like simulated annealing (Hwang, 1988) and tabu
search (Glover, 1989)) the concept of neighbourhood.
Search methods based on this latter concept are itera-
tive procedures in which a neighbourhood N(i) is de-
fined for each feasible solution i, and the next solution
j is searched among the solutions in N(i). Hence, the
neighbourhood is a function N : S → 2
S
that assigns
at each solution in the search space S a (non-empty)
subset of S. In our case, every solution is modelled as
an adjacency matrix, where an entry [i, j] is 1 if in the
current solution an arc is present from node i to node
j, and is 0 otherwise.
The sequence of steps of the hill climbing algo-
rithm, for a minimization problem w.r.t. a given ob-
jective function f , are the following:
1. choose an initial solution i in S;
2. find the best solution j in N(i) (i.e., the solution j
such that f ( j) ≤ f (k) for every k in N(i);
3. if f ( j) > f (i), then stop; else set i = j and go to
Step 2.
To counteract the main limitation of hill climbing
(i.e., getting trapped in a local optimum), more ad-
vanced neighbourhood search methods have been de-
fined. The following section presents the Tabu Search
method, a popular and effective optimization tech-
nique that uses the concept of “memory”.
3.2 Tabu Search
As described in the original work of Glover (Glover,
1989), Tabu Search (TS) is a meta-heuristic that
guides a local heuristic search procedure to explore
the solution space beyond local optimality. One of the
main components of this method is the use of an adap-
tive memory, which creates a more flexible search be-
haviour. Memory-based strategies are therefore the
main feature of TS approaches, founded on a quest for
“integrating principles”, by which alternative forms
of memory are appropriately combined with effective
strategies for exploiting them.
Tabus are one of the distinctive elements of TS
when compared to hill climbing or other local search
methods. The main idea in considering tabus is to
prevent cycling when moving away from local optima
through non-improving moves. When this situation
occurs, something needs to be done to prevent the
search from tracing back its steps to where it came
Combining Bayesian Approaches and Evolutionary Techniques for the Inference of Breast Cancer Networks
219

from. This is achieved by declaring tabu (disallow-
ing) moves that reverse the effect of recent moves.
For instance, let us consider a problem where solu-
tions are binary strings of a prefixed length and the
neighbourhood of a solution i consists of the solutions
that can be obtained from i by flipping only one of its
bits. In this scenario, if a solution j has been obtained
from a solution i by changing one bit b, it is possible
to declare a tabu to avoid to flip back the same bit b of
j for some number of iterations (this number is called
the tabu tenure of the move). Tabus are also useful
to help the search move away from previously visited
portions of the search space and, thus, perform more
extensive exploration.
The basic TS algorithm is reported, considering
the minimization of the objective function f , as fol-
lows:
1. randomly select an initial solution i in the search
space S, and set i
∗
= i and k = 0, where i
∗
is the
best solution so far and k the iteration counter;
2. set k = k + 1 and generate the subset V of the ad-
missible neighbourhood solutions of i (i.e., non-
tabu or allowed by aspiration);
3. choose the best j in V and set i = j;
4. if f (i) < f (i
∗
), then set i
∗
= i;
5. update tabu and aspiration conditions;
6. if a stopping condition is met then stop; else go to
Step 2.
Commonly used conditions to end the algorithm
are when the number of iterations (K) is larger than
the maximum number of allowed iterations, or if no
changes to the best solution have been performed in
the last N iterations (as in our tests).
3.3 Genetic Algorithm
Genetic Algorithms (GAs) are a class of computa-
tional models that mimic the process of natural evo-
lution (Goldberg and Holland, 1988). GAs are often
considered as function optimizers although the range
of problems to which genetic algorithms have been
applied is quite broad. Although different variants ex-
ist, most of the methods called “GAs” have at least
the following elements in common: populations of
chromosomes, selection according to a fitness func-
tion, crossover to produce new offspring, and random
mutation of new offspring.
One of the most important issues when using the
GAs to solve an optimization problem is the way to
encode the candidate solutions, that is the individu-
als in the population, and also the genetic operators
(crossover and mutation). Since, this aspect strongly
depends on the specific problem, here we describe
how GAs have been used to build a Bayesian Net-
work. A candidate solution is represented as a string
s of length equal to n
2
, being n the number of nodes
of the network. Each position s[i] can be either 0 or
1, and the information represents the existence of a
connection among node i/n and node i%n, where the
/ operator denotes the integer division, while the %
operators denotes the rest of the division between i
and n. As an example, s[12] = 1 in a network with
10 nodes means that there is a node between node 1
(12/10) and node 2 (12%10). Nodes are numbered
from 0 to n − 1.
To produce admissible solutions (i.e., in our do-
main a network without loops), it is fundamental to
redefine the classical crossover and mutation opera-
tors. More precisely, we developed a simple but effi-
cient method that guarantees that crossover and muta-
tion will produce Bayesian Networks without loops.
To achieve this goal we associated to each solution
two lists, called forward list and backward list. The
two lists maintain, for each node k, the forward links
(i.e., the set of nodes
ˆ
k for which a connection from
k to
ˆ
k exists) and the backward links (i.e., the set of
nodes
ˆ
k for which a connection from
ˆ
k to k exists). By
using these two linked lists it is simple to assess if a
new connection between two nodes can be created. In
detail, let us assume that the algorithm needs to evalu-
ate whether it is possible to add a connection between
nodes k
1
and k
2
(with k
1
being the origin and k
2
the
destination node of the connection). In this scenario,
it is necessary to iteratively scan all the elements in
the backward list of k
1
and check if in their backward
lists k
2
is present. In this case it would be impossi-
ble to create a connection between k
1
and k
2
without
entering a loop in the structure of the network. In the
same way, it is necessary to iteratively scan all the el-
ements in the forward list of k
2
and check if in their
forward lists k
1
is present. Also in this case, the cre-
ation of the connection from k
1
to k
2
will introduce a
loop in the network.
Hence, the proposed crossover operator works as
follows:
1. choose two individuals p
1
and p
2
as parents,
based on tournament selection;
2. select a single crossover point for both the parents;
3. for every locus i before that point set child
1
[i] =
p
1
[i] and child
2
[i] = p
2
[i];
4. for every locus i beyond that point for which
p
1
[i] is equal to p
2
[i], set child
1
[i] = p
1
[i] and
child
2
[i] = p
2
[i];
5. for every locus i beyond that point for which p
1
[i]
is different from p
2
[i], do the following:
ECTA 2016 - 8th International Conference on Evolutionary Computation Theory and Applications
220

• if p
2
[i] = 0, then set child
1
[i] = 0 and set
child
2
[i] = 1 if and only if it is possible to create
a connection between node i/n and node i%n
(set child
2
[i] = 0 in the opposite case);
• if p
1
[i] = 0, then set child
2
[i] = 0 and set
child
1
[i] = 1 if and only if it is possible to create
a connection between node i/n and node i%n
(set child
1
[i] = 0 in the opposite case);
• update the forward and the backward lists.
The mutation operator we proposed works as fol-
lows:
1. for each locus i of an individual p generate a ran-
dom number r from a uniform distribution. If
r ≤ p
m
(where p
m
is the mutation probability) then
select the locus i for mutation;
2. if p[i] = 1, then set p[i] = 0 and update the forward
and backward lists;
3. in the opposite case (p[i] = 0), check if it is pos-
sible to create a connection between node i/n and
node i%n. If the connection does not introduce a
loop set p[i] = 1 and update the data structures,
else p[i] will remain equal to 0.
The genetic operators described above ensure that
the constraint related to the absence of loops is always
satisfied. Moreover, this allows the GA to avoid to
reject a high number of individuals that do not respect
the constraint. This will result in a beneficial effect on
the execution time of the algorithm.
3.4 Regularizators
As already mentioned, we make use of various like-
lihood scores as fitness functions for the inference of
the network. Such scores, namely loglik, AIC, and
BIC, are implemented by using the bnlearn R pack-
age (Scutari, 2009).
Specifically, we first considered the log-likelihood
score (loglik), that is the logarithm of the previously
mentioned likelihood score. Then, as regularized log-
likelihood scores, we used the Akaike Information
Criterion (AIC) (Akaike, 1992) and the Bayesian In-
formation Criterion (BIC) (Schwarz et al., 1978).
To extend this scores in order to model continuous
random variables, we adopt the multivariate Gaussian
implementation of the log-likelihood score (see (Par-
sons, 2011) for a formal definition of the scores and
(Scutari, 2009) for the adopted implementation).
4 RESULTS
To assess the performance of the different approaches
and regularizators, we have considered the HPN-
DREAM breast cancer network inference challenge.
This challenge comprises three sub-challenges, and
we focused on the first one (Sub-challenge 1). This
sub-challenge consists of two distinct parts: the first
one (Sub-challenge 1A) aims at inferring causal sig-
nalling networks using protein time-course data. The
task spanned 32 different contexts, each defined by a
combination of 4 cell lines and 8 stimuli, which fo-
cus on networks with specific genetic and epigenetic
background. Since for these datasets the real net-
work is unknown, beside training data, further data
(not used during the inference) are available to assess
the causal validity of the inferred networks. The sec-
ond part (Sub-challenge 1B) comprises in silico data
task and also focused on causal networks. Anyway,
differently from the former one, the use of a-priori
biology knowledge to design the network is not al-
lowed. Since for this sub-challenge the protein net-
work is known, the evaluation of the achieved results
can be performed by directly comparing the computed
network with the original one.
More in details, the datasets of Sub-challenge 1A
(“real data”) were generated using Reverse Phase Pro-
tein Array (RPPA) quantitative proteomics technol-
ogy. RPPA is a protein array designed as a micro- or
nano-scaled dot-blot platform that allows the simul-
taneous measurement of protein expression levels in
a large number of biological samples in a quantita-
tive manner, when high-quality antibodies are avail-
able (Spurrier et al., 2008). This challenge focuses
on about 45 phosphoproteins (proteins phosphory-
lated at specific sites). Protein abundance may be in-
fluenced by multiple dynamical processes operating
over multiple time-scales. This challenge does not
focus on long-term changes over days (e.g. rewiring
of networks due to epigenetic changes brought about
by perturbation), hence data comprises protein time-
course data up to 4 hours after ligand stimulation.
Time-course data were acquired under 8 ligand stim-
uli and inhibition of network nodes by one of 3 in-
hibitors plus the vehicle control (cells were serum-
starved and pre-treated with inhibitor prior to lig-
and stimulation). The experiment was carried out
on 4 breast cancer cell lines (namely, BT20, BT549,
MCF7, and UACC812), with abundance of the ∼
45 phosphoproteins measured at 7 time points post-
stimulus. Data are normalized protein abundance
measurements on a linear scale. Table 2 shows the 32
different processed datasets, obtained by each combi-
nation of cell/stimulus, and their compositions, which
are the expression levels of the considered phospho-
proteins with 4 different inhibitors at 7 consecutive
time points.
On the other hand, the in silico challenge aims
Combining Bayesian Approaches and Evolutionary Techniques for the Inference of Breast Cancer Networks
221
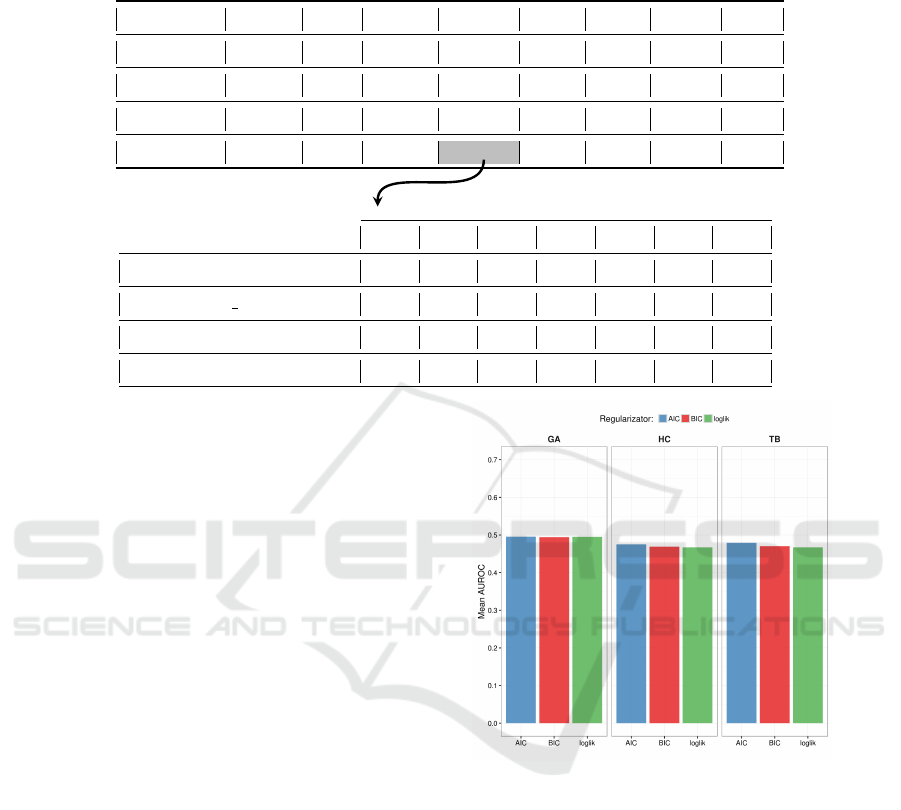
Table 2: The upper table highlights the 32 combinations of cells/stimuli which constitute the processed “real datasets”. The
lower table represents the composition of a single dataset (UACC812/Insulin in the example), which contains the expression
levels of the phosphoproteins with 4 inhibitors at 7 different time points.
Serum PBS NRG1 Insulin IGF1 HGF FGF1 EGF
BT20
BT549
MCF7
UACC812
0m 5m 15m 30m 1h 2h 4h
GSK690693
GSK690693 GSK1120212
PD173074
DMSO
to mimic the key aspects of the RPPA experimental
set up and the characteristics of the proteomic data,
but using a state-of-the-art dynamical model of sig-
nalling. This allows the assessment of inferred net-
works and predicted trajectories against a true gold
standard. A computational signalling model was used
to generate time-courses of phosphoprotein abun-
dance levels. The model describes the biochemistry
underlying a realistic signalling network. Data were
generated for combinations of 2 ligand stimuli (each
one at 2 concentrations, denoted to as “lo” and “hi”)
and 3 inhibitors, or no inhibitor (as for the experi-
mental data described above, cells were pre-incubated
with the inhibitor prior to ligand stimulation). For
each condition, a time-course of 20 phosphoprotein
levels is provided at 10 time points post-stimulus. It
must be noticed that phosphoprotein names have been
anonymized so that detailed prior information from
canonical signalling pathways cannot be used. Efforts
have been made to model the antibody-based readout
of the RPPA platform and its technical variability in
a faithful manner. Three technical replicates are pro-
vided per condition. Data provided to participants are
protein abundance measurements on a linear scale. In
this task, a single network should be inferred in con-
trast to the proteomic data challenge that requires 32
networks.
Following the approach used to evaluate the re-
sults submitted to the challenge, we have considered
the same method to assess the performance of our
predictions. More precisely, in real data, for any
given context, the set of nodes that showed salient
changes under a test inhibitor (here an mTOR in-
hibitor) relative to the control was identified. These
Figure 1: Mean results on the 32 experimental datasets for
the considered approaches.
“gold-standard” sets are derived from (held-out) ex-
perimental data and should not be regarded as repre-
senting a fully definitive ground truth. For each pre-
dicted network, the set of mTOR descendants is pre-
dicted and compared against the experimental one to
obtain the area under the receiver operating charac-
teristic curve (AUROC) score (Hill et al., 2016). Re-
sults are ranked in each of the 32 contexts by AUROC
score, and the mean rank across contexts was used to
provide an overall score and a final ranking. For the
in silico data task, the true causal network was known
and it was used to obtain an AUROC score for each
predicted network. This score has been considered to
determine the final ranking.
By analysing the mean AUROC values computed
on the predictions on the 32 real datasets, which are
ECTA 2016 - 8th International Conference on Evolutionary Computation Theory and Applications
222
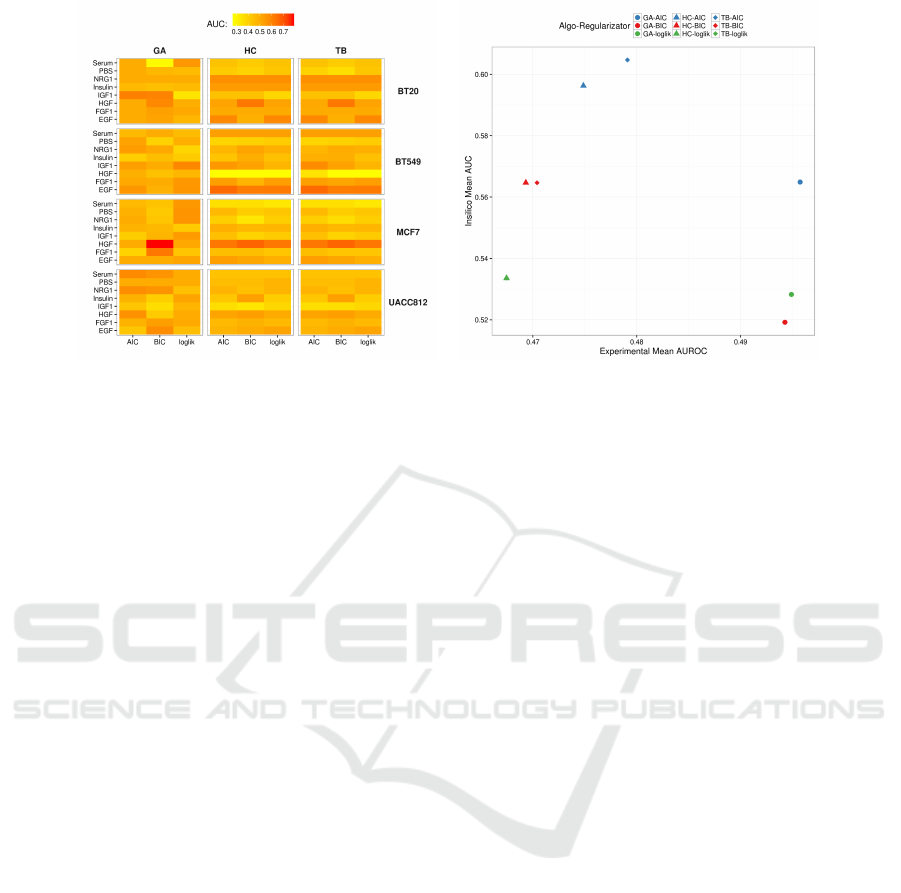
(a) (b)
Figure 2: (a)Heatmap showing the AUC values obtained with each combination of heuristic search method and regulariza-
tor on the 32 experimental datasets. (b) Scores in experimental and in silico data tasks. Each combination of shape/color
corresponds to a specific algorithm/regularizator pair.
reported as bars in the plots in Figure 1, it is possible
to observe that all the tested approaches have similar
performance, with mean values around 0.5.
Anyway, when looking more in details on each
of the 32 datasets, we can draw more accurate con-
siderations about the behaviour of the tested tech-
niques. In particular, as showed in the heatmap in
Figure 2(a), on the processed datasets we have ob-
tained AUROC values ranging from 0.3 to 0.7. As
corroborated by several studies present in literature,
these results highlight the fact that HC (hill climbing)
and TB (tabu search) have almost the same behaviour,
also, w.r.t. the considered regularizator, on the major-
ity of the datasets. On the other hand, GA (genetic al-
gorithm) presents slightly different results than those
obtained by the other two methods and, moreover, it
seems that the results are affected by the considered
regularizator. Interestingly, when looking at the in sil-
ico AUC values, we can observe that, for each reg-
ularizator, HC and TB perform better on the in sil-
ico dataset, while GA is slightly worse; the opposite
situation is observed in the real datasets, where the
latter method (i.e. genetic algorithms) achieves bet-
ter results with respect to the two former techniques
(i.e., hill climbing and tabu search). The scatter plot
in Figure 2(b) shows a comparison of the mean AUC
results on the in silico dataset against the AUROC
mean values on the real datasets obtained with all the
employed approaches.
To assess the quality of the obtained results, we
performed a comparison with those obtained by the
participants of the challenge. More precisely, as re-
ported in (Hill et al., 2016), several different tech-
niques have been used to reconstruct the network pro-
posed in this challenge, which can be distinguished
based on the fact that a prior knowledge has been em-
ployed in order to improve the predictions, and also
based on the reconstruction method (Bayesian net-
works in our case). From the results on the in sil-
ico dataset, ranked by the mean AUC, we observed
that our best performer (TB with AIC) obtained a
value of 0.6, which is better than all the other meth-
ods based on Bayesian networks and ranks in the top
15% of the overall evaluated techniques. On the other
hand, on the 32 real datasets our results are similar
to those obtained by methods based on Bayesian net-
works, which present values around 0.5. Both these
results are not surprising, since we do not use any
prior knowledge on the input data (resulting in good
performance on the in silico dataset), and also the
number of observations in each of the 32 real datasets
is quite low compared to the number of nodes (phos-
phoprotein) of the networks to reconstruct, hence pe-
nalizing Bayesian approaches, making the inference
task difficult.
5 CONCLUSIONS
In this work, we studied the inference of causal
molecular networks, specifically focusing on signal-
ing downstream of receptor tyrosine kinases. We
modeled relationships (edges) in causal molecular
networks (’causal edges’) as directed links between
nodes, in which inhibition of the parent node can lead
to a change in the abundance of the child node, either
by direct interaction or via unmeasured intermediate
nodes.
Combining Bayesian Approaches and Evolutionary Techniques for the Inference of Breast Cancer Networks
223

To this extent, we have tested different methods
to reconstruct (Bayesian) networks on real and in sil-
ico datasets proposed in the HPN-DREAM challenge.
Specifically, we analyzed the performance of different
optimization search schemes, i.e., Hill climbing (HC),
Tabu seach (TS) and Genetic algorithms (GA), and
various likelihood scores, i.e., loglik, AIC and BIC.
This analysis seems to show a better performance of
more sophisticated search strategies like GA on real
datasets, even if on in silico data it is shown that eas-
ier search schemes as HC and TS also prove to be very
effective.
Furthermore, we find the obtained results to be en-
couraging, especially considering the fact the we have
employed “standard” versions of the algorithms for
the reconstruction of the network without making use
of any biological prior.
REFERENCES
Akaike, H. (1992). Information theory and an extension of
the maximum likelihood principle. In Breakthroughs
in statistics, pages 610–624. Springer.
Akutsu, T., Miyano, S., Kuhara, S., et al. (1999). Identi-
fication of genetic networks from a small number of
gene expression patterns under the boolean network
model. In Pacific symposium on biocomputing, vol-
ume 4, pages 17–28. Citeseer.
Bansal, M., Belcastro, V., Ambesi-Impiombato, A., and
Di Bernardo, D. (2007). How to infer gene networks
from expression profiles. Molecular systems biology,
3(1):78.
Friedman, N., Linial, M., Nachman, I., and Pe’er, D. (2000).
Using bayesian networks to analyze expression data.
Journal of computational biology, 7(3-4):601–620.
Glover, F. (1989). Tabu search-part i. ORSA Journal on
computing, 1(3):190–206.
Goldberg, D. E. and Holland, J. H. (1988). Genetic al-
gorithms and machine learning. Machine learning,
3(2):95–99.
Hill, S. M., Heiser, L. M., Cokelaer, T., Unger, M., Nesser,
N. K., Carlin, D. E., Zhang, Y., Sokolov, A., Paull,
E. O., Wong, C. K., et al. (2016). Inferring causal
molecular networks: empirical assessment through a
community-based effort. Nature methods, 13(4):310–
318.
Hwang, C.-R. (1988). Simulated annealing: theory
and applications. Acta Applicandae Mathematicae,
12(1):108–111.
Imoto, S., Goto, T., Miyano, S., et al. (2001). Estimation
of genetic networks and functional structures between
genes by using bayesian networks and nonparametric
regression. In Pacific symposium on Biocomputing,
volume 7, pages 175–186.
Kabir, M., Noman, N., and Iba, H. (2010). Reverse engi-
neering gene regulatory network from microarray data
using linear time-variant model. BMC bioinformatics,
11(1):1.
Parsons, S. (2011). Probabilistic graphical models: Prin-
ciples and techniques. The Knowledge Engineering
Review, 26(02):237–238.
Pearl, J. (2003). Causality: models, reasoning and infer-
ence. Econometric Theory, 19:675–685.
Schramm, G., Kannabiran, N., and K
¨
onig, R. (2010). Reg-
ulation patterns in signaling networks of cancer. BMC
systems biology, 4(1):1.
Schwarz, G. et al. (1978). Estimating the dimension of a
model. The annals of statistics, 6(2):461–464.
Scutari, M. (2009). Learning bayesian networks with the
bnlearn r package. arXiv preprint arXiv:0908.3817.
Spurrier, B., Ramalingam, S., and Nishizuka, S. (2008).
Reverse-phase protein lysate microarrays for cell sig-
naling analysis. Nature protocols, 3(11):1796–1808.
Stolovitzky, G., Monroe, D., and Califano, A. (2007). Di-
alogue on reverse-engineering assessment and meth-
ods. Annals of the New York Academy of Sciences,
1115(1):1–22.
Zou, M. and Conzen, S. D. (2005). A new dynamic
bayesian network (dbn) approach for identifying gene
regulatory networks from time course microarray
data. Bioinformatics, 21(1):71–79.
ECTA 2016 - 8th International Conference on Evolutionary Computation Theory and Applications
224
