
On the Relationship between a Computational Natural Logic and
Natural Language
Troels Andreasen
1
, Henrik Bulskov
1
, Jørgen Fischer Nilsson
2
and Per Anker Jensen
3
1
Computer Science, Roskilde University, Roskilde, Denmark
2
Mathematics and Computer Science, Technical University of Denmark, Lyngby, Denmark
3
International Business Communication, Copenhagen Business School, Frederiksberg, Denmark
Keywords:
Computational Semantics of Natural Languages, Models of Computation for Natural Language Processing,
Bio-Information and Natural Language.
Abstract:
This paper makes a case for adopting appropriate forms of natural logic as target language for computa-
tional reasoning with descriptive natural language. Natural logics are stylized fragments of natural language
where reasoning can be conducted directly by natural reasoning rules reflecting intuitive reasoning in natural
language. The approach taken in this paper is to extend natural logic stepwise with a view to covering succes-
sively larger parts of natural language. We envisage applications for computational querying and reasoning, in
particular within the life-sciences.
For better or for worse, most of the reasoning that is done in the
world is done in natural language.
G. Lakoff: Linguistics and Natural Logic, Synthese, 22, 1970.
1 INTRODUCTION
Traditionally, computational reasoning with informa-
tion given in natural language is carried out by con-
ducting a translation of sentences into first order
predicate logic, see e.g. (Fuchs et al., 2008; Kuhn
et al., 2006), or derivatives thereof such as a descrip-
tion logic dialect, as in (de Azevedo et al., 2014;
Thorne et al., 2014). There are also natural logic
approaches which extend syllogistic proof systems
(Pratt-Hartmann and Moss, 2009) or which call on
forms of logical type theory, thereby taking advan-
tage of an assumed compositional semantics for natu-
ral language drawing on higher order denotations (Fy-
odorov et al., 2003).
The project described here relies on appropri-
ate forms of natural logic decomposed into graph-
structured knowledge bases. Natural logics are tiny,
stylized fragments of natural language in which the
deductive logical reasoning can be carried out directly
by simple, intuitive rules, that is, without taking resort
to predicate-logical reasoning systems such as reso-
lution. Natural logic originates from the traditional
Aristotelian categorial syllogistic logic (van Benthem,
1986; Kl
´
ıma, 2010; Nilsson, 2015), which became
further developed and refined in late medieval times.
However, in the course of the late 19th century devel-
opment, forms of logic coming close to natural lan-
guage were largely deemed obsolete by Frege’s intro-
duction of the more general, mathematically inclined
quantifier-based predicate logic. As is well-known,
the latter subsequently prevailed throughout the 20th
century.
This paper pursues the idea of choosing natural
logic as a target language for dealing computation-
ally with appropriately constrained and regimented,
yet rich, forms of affirmative sentences in natural lan-
guage. The purported methodological advantage of
the present approach lies in the proximity of natu-
ral logic to natural language, very much in contrast
to predicate logic. Indeed then, the translation of
the considered natural language fragments becomes
a partial recasting of the considered sentences into an
even smaller language fragment, namely natural logic
sentences. Obviously, the chosen natural logic then
determines and confines the semantic range for partial
coverage of the considered sentences. A related use of
natural logic principles for partial computational un-
derstanding of natural language is found in (MacCart-
ney and Manning, 2009). The language translational
relationships are planned to be initially implemented
using the well-known, rather unsophisticated definite
Andreasen, T., Bulskov, H., Nilsson, J. and Jensen, P.
On the Relationship between a Computational Natural Logic and Natural Language.
DOI: 10.5220/0005848103350342
In Proceedings of the 8th International Conference on Agents and Artificial Intelligence (ICAART 2016) - Volume 1, pages 335-342
ISBN: 978-989-758-172-4
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
335

clauses (clause grammars).
The paper is structured as follows: In section 2
we describe the semantic basis of the considered di-
alect of natural logic presented in section 3. In section
4 we discuss various mainly conservative extensions
of the natural logic in order to accommodate expres-
sion forms common in natural language. Section 5
discusses various sentence cases and the problems in-
volved in approaching free natural language formula-
tions. Finally, in section 6 a summary concludes the
paper.
2 SEMANTIC MOTIVATION
Our semantic framework comprises a selection of
stated classes of entities together with binary relation-
ships between the classes akin to the popular entity-
relationship models. First of all, there is a funda-
mental isa subclass relationship known from formal
ontologies. In addition, class-class relationships may
be be introduced according to needs, as discussed in
(Smith et al., 2005; Schulz and Hahn, 2004; Bittner
and Donnelly, 2007; Yu, 2006).
It is a key feature of our approach that the given
named classes may be used to form subclasses ad li-
bitum by restriction with relationships to other classes
in the natural logic. As an example, given the classes
cell and hormone and the relation(transitive verb) pro-
duce, one may form the subclass
cell that produce hormone
This is a phrase in the applied natural logic forming a
new class which is a subclass of cell.
Unlike what is the case in predicate logic, in our
framework the entities belonging to the classes are not
dealt with explicitly. Individual entities may, how-
ever, if necessary, be dealt with as stipulated singleton
classes (having no subclasses in so far as the empty
class is left out in our setup).
This basis, while being application-neutral at the
outset, appears to be particularly useful for applica-
tions within the bio-sciences as discussed in (Smith
et al., 2005; Schulz and Hahn, 2004) as well as in
our (Andreasen et al., 2014a; Andreasen et al., 2014b;
Andreasen and Nilsson, 2014; Nilsson, 2015; An-
dreasen et al., 2015). General, natural language de-
scriptions in natural sciences abound with classes, let
us just mention the Linnean and chemical and medical
taxonomies. The class-relationship framework might
also find more innovative use for ontology-structured
knowledge base concept organization and specifica-
tion in semi-exact sciences, e.g. in linguistics.
3 CORE NATURAL LOGIC
Having introduced the semantic motivation, we turn
next to the logical sentences serving the mentioned
class-relationship setup. We consider a natural logic
which has the general form of expression shown in
(1):
Q
1
Cterm
0
R Q
2
Cterm
00
(1)
where
• Q
i
are quantifiers (determiners) every/all, some/a,
• the grammatical subject term Cterm
0
and the
grammatical object term Cterm
00
are class expres-
sions, and
• R is a relation name.
In linguistic parlance, the Cterms are noun
phrases, and R is a transitive verb. In simpler cases
Cterms are just class names (nouns) C. Among the
quantifier options here, we focus on the quantifier
structure
every Cterm
0
R some Cterm
00
(2)
This form is pivotal in our treatment because it repre-
sents the default interpretation of sentences like be-
tacells produce insulin in ordinary descriptive lan-
guage. Further, it conforms with the functioning of
the class restrictions as to be described. Example:
every betacell produce some insulin. An explication
of this form in predicate logic with quantifier struc-
ture ∀x∃y is straightforward, cf. our references above,
and therefore not repeated here. The inherent linguis-
tic structural ambiguity corresponding to the scope
choice ∀x∃y versus ∃y∀x is overcome by stipulating
the ∀x∃y reading, which is the useful one in practice.
See also (2) Accordingly, we have
∀x(betacell(x) → ∃y(produce(x,y) ∧ insulin(y))
Observe that we are not going to translate the sen-
tences into their predicate logical form with individ-
ual variables. Rather, it is a crucial feature of our ap-
proach that we decompose the sentences into simpler
constituents forming a graph without variables as ex-
plained in section 3.4.
Our natural logic notations rely on the convention
that if no quantifiers are mentioned explicitly, the in-
terpretation follows the scope pattern ∀x∃y. Thus,
the sentence betacell produce insulin is semantically
equivalent to our sample sentence above, every beta-
cell produce some insulin. Note further that we per-
sistently use uninflected forms of nouns and verbs
rather than morphologically correct forms in our nat-
ural logic expressions.
PUaNLP 2016 - Special Session on Partiality, Underspecification, and Natural Language Processing
336

3.1 Subclass Through Copula Sentence
Within the above natural logic affirmative sentence
template in (1), there is an extremely important sub-
class, namely the copula sentence form shown in (3)
Cterm
0
isa Cterm
00
(3)
known from categorial syllogistic logic. cf. (Nils-
son, 2013). Using the symbol C in class names as in
C
0
isa C
00
we say that the class C
0
specializes the class
C
00
, and, conversely, that C
00
generalizes the class C
0
.
As explained in (Nilsson, 2013) the copula form (3)
may actually be understood as a special case of the
above general form (2) with the relation being equal-
ity. For example, the sentence betacell isa cell is pred-
icate logically construed as
∀x(betacell(x) → ∃y(x = y ∧ cell(y))
giving in turn
∀x(betacell(x) → cell(x))
Note once again that we are not using these predicate
logical forms in our reasoning with natural logic.
The categorial syllogistic no C
0
isa C
00
is not made
available, since class disjointness is assumed initially
for pairs of classes by default in our setup, cf. (Nils-
son, 2015), which also discusses the relation to the
well-known square of opposition in traditional logic.
The upshot of our convention is that two classes are
disjoint unless one is stipulated as a subclass of the
other, or that they have a common subclass introduced
by the copula form. This default convention conforms
with use of classes in scientific practice as reflected in
formal ontologies. However, one may observe that the
convention deviates from the description logic prin-
ciple, which follows predicate logic with the open
world assumption.
3.2 Simple and Compound Class Terms
Compound terms Cterm in the natural logic take the
form of a class name C adorned with various forms of
restrictions giving rise to a virtually unlimited num-
ber subclasses of C. Linguistically, this “generativity”
is provided by constructions like restrictive relative
clauses and adnominal prepositional phrases (PPs).
Accordingly, in the present context we consider
Cterm in the form of a class name (noun) C option-
ally followed by
• a stylized relative clause: that R Cterm
or optionally by
• a PP in the logical form R
prep
Cterm, in turn
optionally followed by a relative clause.
The relation R
prep
is to be provided by the entry for
the pertinent preposition in the applied vocabulary.
Sample class terms illustrating these patterns:
cell
cell that produce hormone
cell in pancreas
cell in pancreas that produce hormone
Ontologically, these four classes form a (trans-
hierarchical) diamond by the isa subclass relation:
cell-in-pancreas-that-produce-hormone
cell-that-produce-hormone cell-in-pancreas
cell
The sample class term cell in pancreas that pro-
duce hormone is aligned as in cell (in pancreas) (that
produce hormone).
For the moment we disregard the more tricky re-
strictions provided by adjectives (even when assumed
to behave restrictively), noun-noun compounds
1
, and
genitives. This is because these constructs, unlike
the case of relative clauses and PPs, do not explic-
itly yield a specific relation R, cf. the discussions in
(Jensen and Nilsson, 2006; Vikner and Jensen, 2002).
The natural logic sub-language where Cterm is
simply a class name we call atomic natural logic.
As described below, in due course we shall also
admit conjoined constructions with the conjunction
and in class terms, and, further, compound relation
terms, Rterm for R, linguistically comprising selected
adverbs and adverbial PPs modifying verbs and verb
phrases. See also (Andreasen et al., 2014b) for vari-
ous extensions of the natural logic.
3.3 Reasoning with Natural Logic
The key inference rules for the considered natural
logic are the so-called monotonicity rules (van Ben-
them, 1986). They very intuitively admit specializa-
tion of the grammatical subject class and generaliza-
tion of the grammatical object class. Accordingly,
given A isa B and [every ] B R [some] C, one may de-
rive [every] A R [some] C,
A isa B [every] B R [some] C
[every] A R [some] C
and given [every] B R [some] C and C isa D, one may
1
Some class names (given terms) in an application may
consist of more than one word,but are still to be considered
as simple, fixed terms.
On the Relationship between a Computational Natural Logic and Natural Language
337
insulin
hormone
cell-that-
produce-hormone
produce
cell
cell-that-
produce-insulin
produce
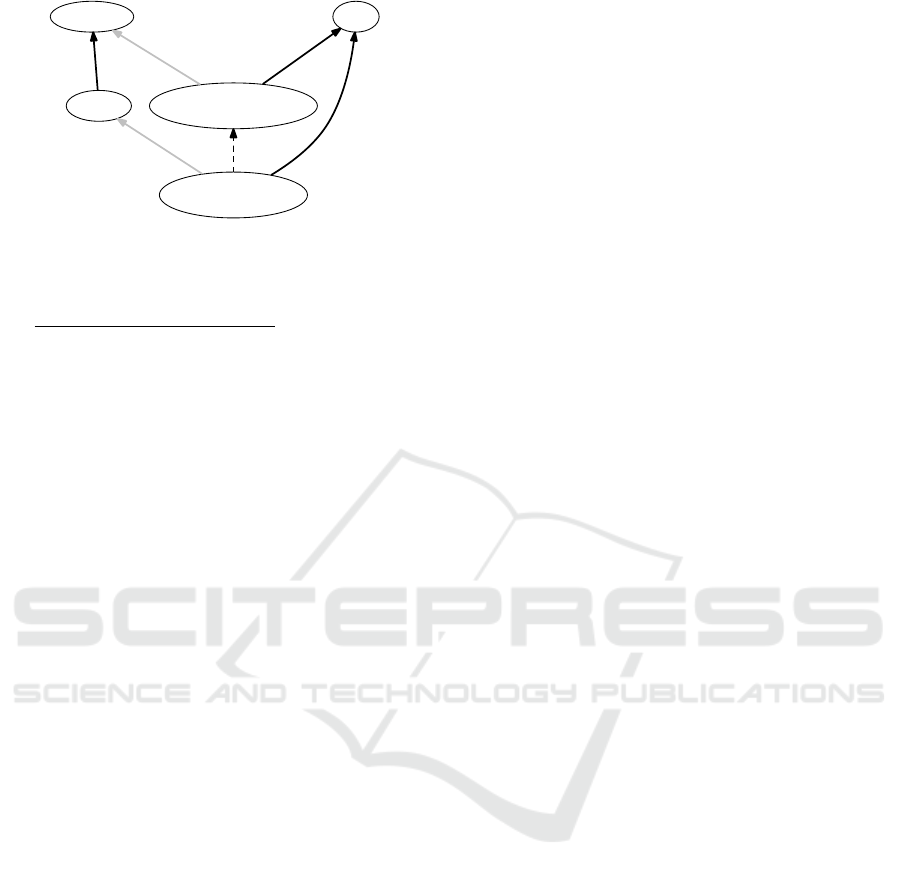
Figure 1: Inferred inclusion by subsumption.
derive [every] B R [some] D,
[every] B R [some] C C isa D
[every] B R [some] D
In particular, these rules provide transitivity of the isa
subclass relationship with R being isa.
In addition, we provide a subsumption inference
rule which makes the properties assigned to classes
act restrictively as detailed with an algorithm in (An-
dreasen et al., 2015). By way of example, the sub-
sumption rule ensures that
cell that produce insulin
is recognized as a specialization of
cell that produce hormone,
given that insulin isa hormone, cf. figure 1.
3.4 Atomic Natural Logic as Graph
In our framework and prototype system, the core
natural logic introduced above is decomposed into
atomic natural logic devoid of compound class names,
that is, Cterm is simply a class name C. This is ac-
complished by introduction of fresh, internal class
names such as cell-that-produce-hormone, which is
formally conceived of as a class name. In turn, this
is defined by two atomic natural logic sentences
cell-that-produce-hormone isa cell
cell-that-produce-hormone produce hor mone.
The knowledge base of the decomposed sentences
may be viewed as one single labeled graph whose
nodes are uniquely labeled with classes. Except for
those relationships that follow from transitivity, we
make sure that all valid isa relationships between
nodes are materialized in the graph by the subsump-
tion rule, so that for instance
cell-that-produce-insulin isa
cell-that-produce-hormone
is recorded. As such, the graph appears as an
extended formal ontology with the isa relationship
forming the skeleton, as it were.
In our system, besides deductive querying the
graph is used for pathfinding between classes (An-
dreasen et al., 2015). Actually, in our system the
atomic natural logic graph is embedded in function-
free logical clauses (e.g. DATALOG). This embedding
approach means that natural logic sentences become
encoded as variable-free logical terms. The logical
variables in the clauses then range over class and re-
lationship terms enabling formulation of the inference
rules and hence reasoning and deductive querying.
The clausal embedding also facilitates formulation of
domain specific inference rules such as transitivity of
causation (Andreasen et al., 2014b).
4 EXTENDING CORE NATURAL
LOGIC
We now turn to a partial treatment of natural language
sentences using natural logic as a vantage point. In
a conventional approach, one may proceed by devis-
ing a (partial) translation from the considered natural
language text sentence by sentence into natural logic.
Here, we choose to proceed by introducing a series of
conservative extensions to core natural logic. Thus,
these extensions do not increase the semantic range
of the core natural logic as stipulated above. How-
ever, the extensions provide paraphrases commonly
encountered in natural language. These extensions,
when taking jointly, form an extended natural logic
coming closer to free natural language formulations,
remaining, however, within the confines of the seman-
tics of core natural logic. The extensions may be used
in turn in developing a partial translator from natural
language into core natural logic.
4.1 Extension with Conjunctions
Let us first consider conjunctions of Cterms including
the linguistic conjunction and assuming distributive
(in contrast to collective) readings. Conjunctions in
the grammatical object
Cterm
0
R Cterm
00
1
and Cterm
00
2
as in
pancreas contain betacell and alphacell
straightforwardly give rise to the decomposition:
Cterm
0
R Cterm
00
1
Cterm
0
R Cterm
00
2
Conjunctions in the grammatical subject
Cterm
0
1
and Cterm
0
2
R Cterm
00
as in
betacell and alphacell is-contained-in pancreas
are conventionally interpreted as disjunction rather
than overlap of the two classes and therefore decom-
posed into
Cterm
0
1
R Cterm
00
Cterm
0
2
R Cterm
00
PUaNLP 2016 - Special Session on Partiality, Underspecification, and Natural Language Processing
338

These conventions are justified by the underlying
predicate logical explication of core natural logic.
The linguistic disjunction or in the linguistic sub-
ject seems irrelevant from the point of view of the
considered domains. It may be considered a case
where predicate logic covers more than needed.
More interesting are disjunctions in the grammat-
ical object, viz.
Cterm
0
R Cterm
00
1
or Cterm
00
2
which cannot simply be decomposed into two core
natural logic sentences. One approach is to appeal
to a common general term Cterm
sup
for Cterm
00
1
and
Cterm
00
2
if one is available in the KB. More precisely,
one seeks a Cterm
sup
, such that
Cterm
00
1
isa Cterm
sup
Cterm
00
2
isa Cterm
sup
and such that for all different Cterm
x
having these
properties Cterm
sup
isa Cterm
x
. This supremum re-
quirement seems reasonable in cases where the con-
sidered disjunction is pragmatically relevant at all.
Notice that all of the above reductions of conjunc-
tions endorse the desired commutativity and associa-
tivity properties.
It goes without saying that the presence of con-
junctions together with relative clauses and preposi-
tions gives rise to structural ambiguities. Introduction
of appropriate default readings and/or addition of aux-
iliary parentheses are the simplest ways to eliminate
these.
Collective, i.e. non-distributive, readings such as
co-presence of A and B cause C call for separate
treatment, which goes beyond the scope of the present
approach.
4.2 Extension with Appositions and
Parenthetical Relative Clauses
Let us consider natural logic sentences
Cterm
0
R Cterm
00
extended with appositions bounded by commas
Cterm
0
, [a|an] Cterm
appo
, R Cterm
00
This is paraphrased into the pair
Cterm
0
R Cterm
00
Cterm
0
isa Cterm
appo
Analogously for the grammatical object, Cterm
00
, as
in
betacell produce insulin, a peptide hormone
In this extended natural logic, the pronoun ’that’
is formally set off for restrictive relative clauses as
accounted for above.
By contrast, consider the case of parenthetical rel-
ative clauses for which in the formal natural logic we
use ’which’ together with commas
Cterm
0
, which R
par
Cterm
par
, R Cterm
00
as in
insulin, which isa peptide hormone, ...
Retaining logical equivalence, this can be para-
phrased into the joint pair
Cterm
0
R Cterm
00
Cterm
0
R
par
Cterm
par
and similarly and recursively for Cterm
00
and
Cterm
par
.
4.3 Beyond Core Natural Logic
From the point of view of application functionalities,
verbs should be allowed extensions with adverbial
PPs yielding restricted relations, Rterm, for plain R,
say, as in
A produce in pancreas B
with variants A produce B in pancreas and in pan-
creas A produce B, with obvious additional structural
ambiguity problems.
On our agenda for non-conservative extensions of
core natural logic, let us mention passive voice verb
forms, nominalisation, and plural formation. As far
as negation is concerned we rely throughout on the
closed world assumption in the query answering.
Of course there are numerous genuine (that is,
non-conservative) language extensions which go be-
yond the semantic range of core natural logic, even
within the given scope of monadic and dyadic re-
lations in affirmative sentences. Thus admission of
anaphora as in the infamous donkey sentences breaks
the boundaries as mentioned in (Kl
´
ıma, 2010). An ex-
ample of this is seen in every cell that has a nucleus
is-controlled-by it. The point is that in the applied nat-
ural logic, the subject noun term and the object noun
term are independent, connected solely by the relator
verb and unconnected by anaphora.
5 COMPUTING NATURAL
LOGIC FROM SENTENCES
Methodologically, rather than the usual forward or
bottom-up translation following the phrase structure,
we devise a top down processing governed by the nat-
ural logic. In this process we try to cover as much as
possible of the considered sentence in a (partial) “best
fit” process.
A prototype is under development and is currently
functioning in a preliminary version. In the present
approach natural language input is processed sentence
by sentence. Thus sentential context is not exploited.
Each input sentence is preprocessed for markup, and
the result is further processed by a parser that also
On the Relationship between a Computational Natural Logic and Natural Language
339

insulin
peptide_hormone-producedby-
betacell-in-pancreas
betacell-in-pancreas
producedby
peptide_hormone
pancreas
in
betacell
Figure 2: Graph representation of the sentence insulin is a peptide hormone produced by betacells in the pancreas.
functions as a natural logic generator. During the pre-
processing the sentence is tokenised into a list of lists,
where each word from the sentence is represented
by a list of possible lexemes specifying base form
of word and word category (part of speech) for each
word. A lexeme is included as possible if it matches
the lemma of the input form of the word. Finally, the
preprocessing applies a domain specific vocabulary to
identify multiword expressions in the input sentence.
To ensure that multiwords are treated as inseparable
units they are replaced by unique symbols. A prepro-
cessing of the sentence: Insulin is a peptide hormone
produced by betacells in the pancreas returns the fol-
lowing tagged and lemmatised word list, where the
word sequences ’is a’, ’peptide hormone’ and ’pro-
duced by’ are replaced by symbols:
{{insulin/NN}, {isa/VB}, {peptide hormone/NN},
{produced by/VB}, {betacell/NN}, {in/JJ, in/NN,
in/RB, in/IN}, {pancreas/NN}}
Each possible lexeme for a word (and multiword) W
i
is included as an element L
i j
/C
i j
if W
i
has lemma L
i j
for category C
i j
. The categories are denoted using
Penn Treebank POS-tags. VB, NN, RB, JJ and IN cor-
respond to verb, noun, adverb, adjective and preposi-
tion, respectively.
Our approach to recognizing and deriving natu-
ral logic expressions from natural language texts can
be considered a knowledge extraction task where the
goal is to extract expressions that cover as much as
possible of the meaning content from the source text.
The search is guided by a natural logic grammar,
and the aim is to create propositions that comprise
well-formed natural logic expressions. The “best fit”
approach is basically a guiding principle aiming for
largest possible coverage of the input text. Thus, if
an expression, that covers the full input sentence can
be derived, it would be considered the “best”, and if
not, the aim is a partial coverage where larger means
“better”.
In the prototype, we apply a simple approach to
deriving a partial coverage. A “best fit” is provided
by iterating through a series of “sub-sentences” of the
input sentence of repeatedly smaller size until one is
found from which a proposition can be derived. A
sub-sentence arises from removing zero, one or more
words from the input sentence. This approach has ob-
vious drawbacks, most importantly, it’s quite ineffi-
cient – especially due to load from forcing the parser
to repeat identical subtasks over and over again. How-
ever, it has one important advantage as a prototype
approach – it allows a clear separation of parsing and
selection of partial expressions.
As far as parsing is concerned, and as already
mentioned, our approach is a top down processing
governed by the natural logic. The word list given as
input is ambiguous due to the multiple categories as-
signed to each word. Thus, the parser should be able
to recognize an input proposition if such exists for at
least one combination of possible lexemes of the in-
put words. Therefore, in addition to processing the
grammar (given below), the parser must ensure that
all combinations are tried before failing the recogni-
tion of a proposition.
Core natural logic, as described in section 3, with-
out the extensions sketched in section 4, can be spec-
ified by the following grammar.
Prop ::= Cterm R Cterm
Cterm ::= NN [RelClauseterm | Prepterm]
RelClauseterm ::= [that|which|who] R Cterm
R ::= VB
R
Prep
::= IN
Prepterm ::= R
Prep
Cterm
Notice that terminals are specified as either specific
words or word categories using Penn Treebank-tags.
Example 1: The preprocessing result of the exam-
ple sentence insulin is a peptide hormone produced
PUaNLP 2016 - Special Session on Partiality, Underspecification, and Natural Language Processing
340
cell-that-
produce-insulin
cell insulin
produce
pancreatic-gland
located:in
Figure 3: Graph representation of the sentence cells that produce insulin are located in the pancreatic gland
by betacells in pancreas is shown above. In this case,
the full sentence can be recognized as a proposition
with the given grammar and the following arcs can be
derived from the parse tree:
insulin isa
peptide hormone-producedby-betacell-in-pancreas
peptide hormone-producedby-betacell-in-pancreas
producedby betacell-in-pancreas
peptide hormone-producedby-betacell-in-pancreas
isa peptide hormone
betacell-in-pancreas in pancreas
betacell-in-pancreas isa betacell
The corresponding graph is shown in graph form in
figure 2.
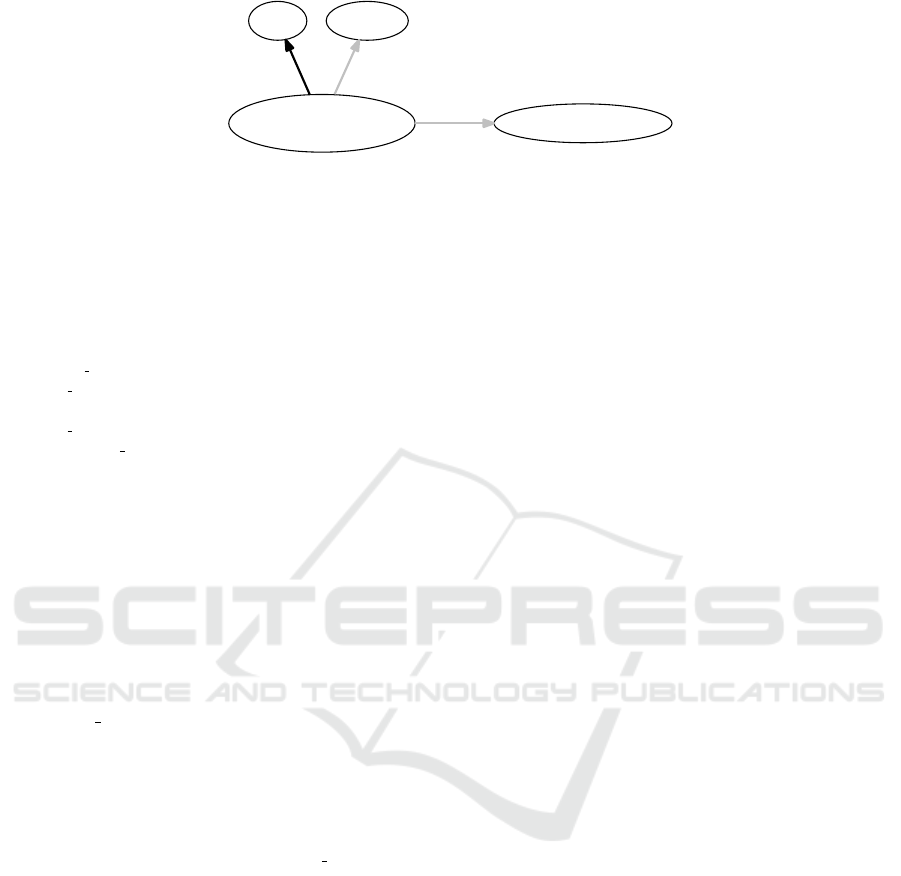
Example 2: Preprocessing the sentence cells that pro-
duce insulin are located in the pancreatic gland leads
to:
{{cell/NN}, {that/WDT}, {produce/VB, pro-
duce/NN}, {insulin/NN}, {locatedin/VB},
{pancreatic gland/NN}}
where ’is located in’ and ’pancreatic gland’ are re-
placed by symbols. As it appears, again the full sen-
tence can be recognized as a proposition. From the
corresponding parse tree the following 3 arcs can be
derived:
cell-produce-insulin locatedin pancreatic gland
cell-produce-insulin produce insulin
cell-produce-insulin isa cell
The result is shown in graph form in figure 3.
Example 3:The grammar above does not cover con-
junctions. Thus pancreas contains betacells and al-
phacells will not be recognized as a proposition.
However, the best-fit principle will iterate through
partial-cover sub-sentences (where one or more words
are left out). Among these are pancreas contain beta-
cell and pancreas contain alphacell, which will both
be recognized. Thus, in this case continued best-
match iteration will lead to a result that complies with
the conjunction.
6 SUMMARY AND CONCLUSION
We have described how to use dedicated forms of nat-
ural logic for partial computational comprehension of
natural language with a particular view to descriptive
scientific corpora within the life sciences. The applied
natural logic, called core natural logic, has a well-
defined semantic foundation in predicate logic, and
we have devised an appropriate inference engine for
query answering functionalities. Core natural logic
is extended in the paper with common paraphrase
schemes.
Our approach features – as explained and illus-
trated with the figures – a shared graph represen-
tation of all sentences. In this representation con-
cepts are (at least ideally) uniquely represented as
nodes. The directed arcs represent atomic natural
logic sentences, and appropriate labelling conventions
ensure that the individual natural logic sentences can
be reconstructed from their atomic components in the
graph, modulo paraphrasing. The graph representa-
tion eases pathfinding between concepts and serves
deductive querying.
Finally, we illustrate processing of sentences from
the text using core natural logic. As a next step, we
are going to try our prototype on selected life science
corpora.
ACKNOWLEDGEMENT
The authors would like to thank the reviewers for their
careful reading of the draft with suggestions for im-
provements and clarifications, and for provision of
further comprehensive references for our continuing
work.
REFERENCES
Andreasen, T., Bulskov, H., Fischer Nilsson, J., and Jensen,
P. A. (2014a). Computing pathways in bio-models de-
rived from bio-science text sources. In Proceedings of
On the Relationship between a Computational Natural Logic and Natural Language
341

the IWBBIO International Work-Conference on Bioin-
formatics and Biomedical Engineering, Granada,
April, pages 217–226.
Andreasen, T., Bulskov, H., Nilsson, J. F., and Jensen, P. A.
(2014b). A system for computing conceptual path-
ways in bio-medical text models. In Foundations of
Intelligent Systems, pages 264–273. Springer.
Andreasen, T., Bulskov, H., Nilsson, J. F., and Jensen, P. A.
(2015). A system for conceptual pathway finding and
deductive querying. In Flexible Query Answering Sys-
tems 2015, pages 461–472. Springer.
Andreasen, T. and Nilsson, J. F. (2014). A case for em-
bedded natural logic for ontological knowledge bases.
In 6th International Conference on Knowledge Engi-
neering and Ontology Development.
Bittner, T. and Donnelly, M. (2007). Logical properties of
foundational relations in bio-ontologies. Artificial In-
telligence in Medicine, 39(3):197–216.
de Azevedo, R. R., Freitas, F., Rocha, R., de Menezes, J.
A. A., and Pereira, L. F. A. (2014). Generating de-
scription logic ALC from text in natural language. In
Foundations of Intelligent Systems, pages 305–314.
Springer.
Fuchs, N. E., Kaljurand, K., and Kuhn, T. (2008). Attempto
controlled english for knowledge representation. In
Reasoning Web, pages 104–124. Springer.
Fyodorov, Y., Winter, Y., and Fyodorov, N. (2003). Order-
based inference in natural logic. Log. J. IGPL,
11(4):385–417. Inference in computational seman-
tics: the Dagstuhl Workshop 2000.
Jensen, P. A. and Nilsson, J. F. (2006). Ontology-based
semantics for prepositions. In Saint-Dizier, P., editor,
Syntax and semantics of prepositions, pages 217–233.
Springer Science & Business Media.
Kl
´
ıma, G. (2010). Natural logic, medieval logic and formal
semantics. Logic, Language, Mathematics, 2.
Kuhn, T., Royer, L., Fuchs, N. E., and Schroeder, M.
(2006). Improving text mining with controlled nat-
ural language: A case study for protein interactions.
In Data Integration in the Life Sciences, pages 66–81.
Springer.
MacCartney, B. and Manning, C. D. (2009). An extended
model of natural logic. In Proceedings of the eighth
international conference on computational semantics,
pages 140–156. Association for Computational Lin-
guistics.
Nilsson, J. F. (2013). Diagrammatic reasoning with classes
and relationships. In Visual Reasoning with Diagrams,
pages 83–100. Springer.
Nilsson, J. F. (2015). In pursuit of natural logics for
ontology-structured knowledge bases. In The Sev-
enth International Conference on Advanced Cognitive
Technologies and Applications.
Pratt-Hartmann, I. and Moss, L. S. (2009). Logics for
the relational syllogistic. Review of Symbolic Logic,
2(4):647–683.
Schulz, S. and Hahn, U. (2004). Ontological foundations of
biological continuants. In Formal ontology in infor-
mation systems. Proceedings of the 3rd international
conference-FOIS, pages 4–6.
Smith, B., Ceusters, W., Klagges, B., K
¨
ohler, J., Kumar, A.,
Lomax, J., Mungall, C., Neuhaus, F., Rector, A. L.,
and Rosse, C. (2005). Relations in biomedical ontolo-
gies. Genome biology, 6(5):R46.
Thorne, C., Bernardi, R., and Calvanese, D. (2014). De-
signing efficient controlled languages for ontologies.
In Bunt, H., Bos, J., and Pulman, S., editors, Com-
puting Meaning, Volume 4, volume 47 of Text, Speech
and Language Technology, pages 149–173. Springer.
van Benthem, J. (1986). Essays in Logical Semantics, Vol-
ume 29 of Studies in Linguistics and Philosophy. D.
Reidel, Dordrecht, Holland.
Vikner, C. and Jensen, P. A. (2002). A semantic analysis of
the english genitive. interaction of lexical and formal
semantics. Studia Linguistica, 56(2):191–226.
Yu, A. C. (2006). Methods in biomedical ontology. Journal
of Biomedical Informatics, 39(3):252 – 266. Biomed-
ical Ontologies.
PUaNLP 2016 - Special Session on Partiality, Underspecification, and Natural Language Processing
342
