Use of GeIS for Early Diagnosis of Alcohol Sensitivity
José Fabián Reyes Román
1,2
and Óscar Pastor López
1
1
Research Center on Software Production Methods (PROS), DSIC, Universitat Politècnica de València,
Camino Vera s/n, 46022, Valencia, Spain
²Department of Engineering Sciences, Universidad Central del Este (UCE), Ave. Francisco Alberto Caamaño Deñó, 21000,
San Pedro de Macorís, Dominican Republic
Keywords: GeIS, SILE, Genomic Diagnostic, Massive Load, Selective Load, Bioinformatics.
Abstract: This study focuses on the importance of Genomic Information Systems (GeIS) today; the results of this
research provide great benefits to the medical community through technological potential. The development
of SILE (Search-Identification-Load-Exploitation) to GeIS improves the databases management with
curated data. The studies are focused on improving the quality of data and time optimization. With SILE we
perform a selective loading of genes and variations found for a specific disease from different data sources
like: NCBI, dbSNP and others. When we worked with a selected group of genes/variations it is possible
guaranteeing a more reliable diagnosis, thus sustaining the increase accuracy of the results with respect to
data quality and improvements over time. Also, we integrate the association of genes/variations with
population studies, for this way providing an early diagnosis for any disease of genetic origin.
1 INTRODUCTION
The development of Genomic Information Systems
(GeIS) brings a great challenge due to what is
known nowadays as genomic chaos, making its
application difficult to genetic tests. Developing and
managing databases for handling this data gives
many benefits to the scientific and technological
community. The area of Bioinformatics requires us
to have a strict control on data manipulation, as it is
based on large amounts of information and data
from different sources, so we must carry out
thorough study that would allow us to ensure
reliable results.
Some of the current data sources, such as NCBI
(Sherry et. al., 2011), OMIM (Hamosh et. al.,) and
Ensembl (Hubbard et. al.,) provide an extensive set
of information, so it is necessary to extract concise
information to help obtaining precise results. That's
why SILE methodology (Search, Identification,
Load, and Exploitation) has come to improve the
tasks of Extraction and Treatment of existing data
sources used.
The Search-Identification-Load-Exploitation
(SILE) is a methodology developed by Óscar Pastor
López within the PROS research group of the
Universitat Politècnica de València, aimed to
improve our process to load the genes and variations
of our Human Genome Database (HGDB).
Its initials are defined as follows:
S
Search - It consists of the exhaustive search of
scientific information (publications, articles, etc.)
To support the genetic association with a specific
disease.
I
Identification - is the process that involves the
medical intervention to provide support in
filtering genes and variations that have greater
incidence in the population.
L
Load - The process of loading the database, is
where we proceed to insert the genes,
chromosomes and variations to the database with
the information treated (curated) and validated,
the load is carried out through various tasks and
data sources.
E
Exploitation - The exploitation is based on the
contents and presentation of the final result.
SILE methodology is performed in various
databases that facilitate search and query of
biomedical information, in this case study we will
use NCBI. The National Center for Biotechnology
Information is a database (library) that collects
information on biomedicine, biotechnology,
biochemistry, genetics, genomics, and genetic
diseases, and others. NCBI also provides some
284
Román, J. and López, Ó.
Use of GeIS for Early Diagnosis of Alcohol Sensitivity.
DOI: 10.5220/0005822902840289
In Proceedings of the 9th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2016) - Volume 3: BIOINFORMATICS, pages 284-289
ISBN: 978-989-758-170-0
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

bioinformatics tools for sequence analysis of DNA,
RNA, and proteins; BLAST is a tool for sequence
alignment of the most used (Sherry et. al., 2001).
NCBI was established on November 4, 1988 in order
to maintain and quickly distribute the amount of
information about molecular biology (NCBI, 2015).
Figure 1: SILE methodology.
1.1 SILE Methodology Applied to
Alcohol Disease
Several studies over the years have supported and
justified the genetic implication of this disease.
Alcoholism is a progressive disease, chronic,
degenerative and genetic, with symptoms that
include a strong need to drink in spite of negative
consequences.
To illustrate these cases, in 2003 "Genetic
alterations related to alcoholism" was published in
the Journal of Neurology (Escarabajal, 2003) until
then, so continued and extensive research was done
on this; for years 2010 and 2011 there were many
contributions validating genes already found, and
showing future projections for new genes that are
under study (Bierut, 2011), (Wang et. al., 2011).
These works present the progress made and confirms
if our genes are related with the alcohol sensitivity
or not. This disease is harmful to the whole body and
especially to the liver, pancreas, heart and the entire
nervous system. Given the importance of the disease
and considering that this can directly affect the
general population, regardless of social status, age or
culture, for this reason we have started to carry out
experiments.
This paper is divided as follows: Section 2
presents the state of the art, which shows the
problems of today within bioinformatics and
manipulation of the big data sources. Section 3
shows the process of the methodology. Section 4
presents the studies to verify (validate) our
hypotheses, in this way we seeing increased quality
of genomic diagnostics. The results of these studies
are presented in Section 5. Finally, Section 6
presents the conclusions and future work.
2 GEIS EVOLUTION (GENOMIC
INFORMATION SYSTEMS)
Currently, the bioinformatics plays an important role
due to the contributions and advances provided to
medicine and technology. In the case of genetic
tests, it has served to provide diagnostic screening to
help preventing and/or treating genetic diseases. The
area of bioinformatics, as well as the study of
genetic diseases associated stays in constant
evolution. It was in 1977 when the DNA sequencing
and the development of software to analyze data
quickly started and for the following year the first
complete gene sequence of an organism was
published (Baxevanis & Ouellette, 2004). There are
countless benefits of genetic testing because they
allow us to identify mutations or alterations in genes,
which is of great use and interest to clinical
medicine, favoring the early diagnosis of diseases
(Villanueva et. al., 2012), (Villanueva Del Pozo,
2011). Similarly, bioinformatics community gives
us: the management of biological databases,
metabolic processes and population genetics,
artificial intelligence, and others (Dawyndt &
Swings, 2006).
Between the bioinformatics branches we can find
researches about: sequence analysis, genome
annotation, analysis of mutations in cancer,
comparative genomics modeling biological systems,
protein-protein coupling, etc. (Baxevanis &
Ouellette, 2004). For the year 2008 there were about
1,200 tests (Reyes, 2013) but they were limited and
expensive, so companies looked for ways to reduce
the cost to facilitate access to people from the
comfort of their homes.
Importantly 23andme is a private U.S. company,
which has been a major driving force in genetic
diagnostics. They have a wide range of services;
they provide information about the genetic history
(related to the ancestors) and personal health (risk of
disease). This data presented is based mostly on
probabilities (Goetz, 2007). Other companies have
been also developed like the case of Genotest
(Reyes, 2013), which provides genetic testing with a
variety of diseases and offers easy access to the
public (Genotest, 2015).
Use of GeIS for Early Diagnosis of Alcohol Sensitivity
285

2.1 Data Manipulation (Genomic
Databases)
Lots of studies have been carried out in the medical
and informatics community, with the aim to find
solutions to the problems of management of
genomic databases, mainly by having to manage
large data sources, so we need to invest more in
time, storage, research, etc. to further improve it.
With current tools and search engines, including:
dbSNP (Sherry et. al., 2011), OMIM (OMIM, 2015),
and others, we are unable to solve certain problems,
but we know we can improve it greatly, because the
use of curated databases allows us to optimize the
performance and report greater precision and
quality. With the execution of the SILE tasks we are
taking a step forward, because we get a better
definition of information, and our main goal is being
better and better. Another issue of discussion is the
relationship between genes (variations, mutations)
and a specific population, where we see how these
are more prevalent in one population or another. By
integrating (insertion) this part within our model of
the database, we will create a custom web services
with great reliability.
3 METHODOLOGY
The methodology used in the experiments are
implemented with SILE for the genomic domain.
3.1 Search
The SILE methodology to beside the medical
progresses (scientific community), and conducting a
series of studies to the alcohol sensitivity, have
allowed us to refer to a group of genes that are
highly associated with the disease.
Figure 2: Search and Identification Process (SILE).
3.2 Identification
Our studies focus on genes ADHIB, ALDH2,
GABRA2, which have validation and large amounts
of evidence and studies in the medical field. These
studies have allowed us to analyse variations as units
which are developed further in one population or
another.
Figure 2 presents the genes in which higher
incidences for alcohol sensitivity were found. In the
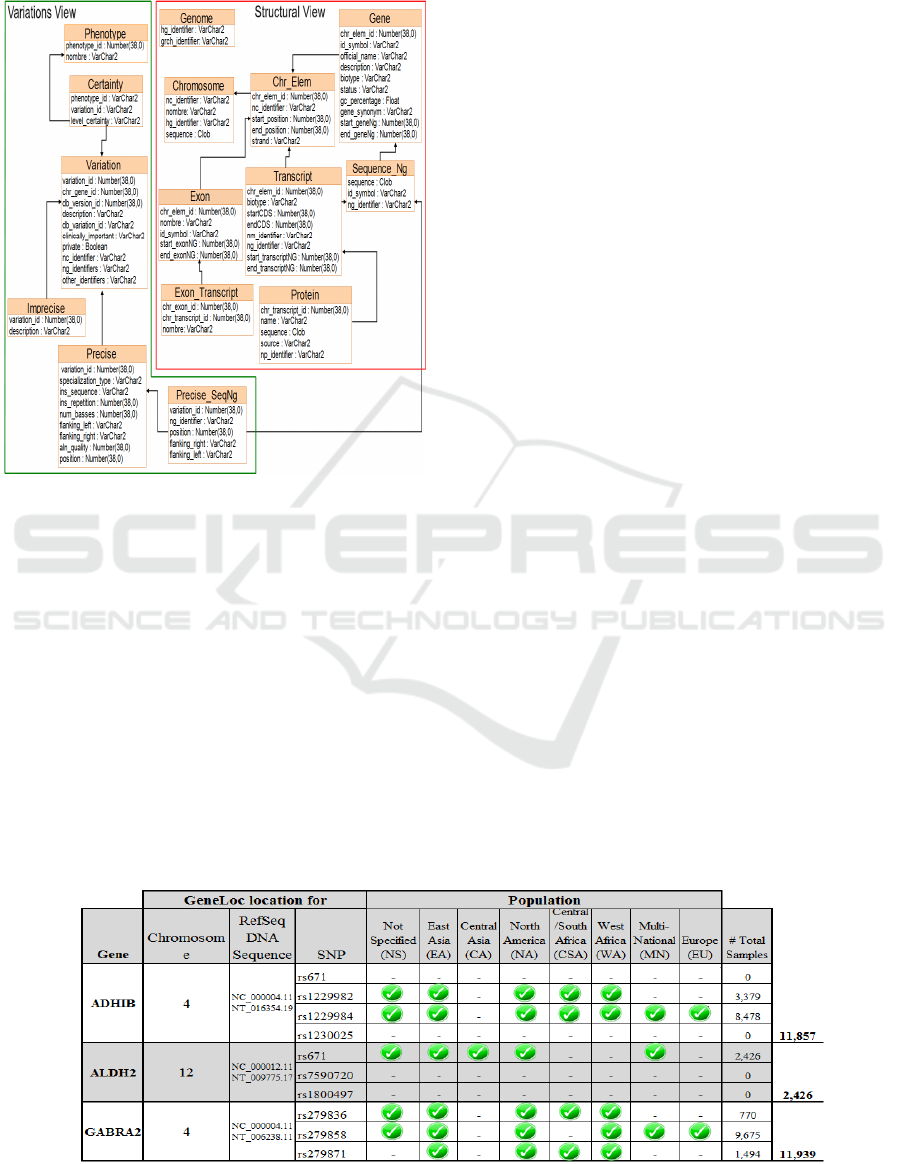
table 2 we can see information about the location of
genes, including a study of the population taken
from GenesCard (Bierut, 2011) as a summary of
NCBI (Wang et. al., 2011) of tests carried out to
different users. When we manage the selective load
(Reyes, 2013), we spent more time in the initial
stages of research and identification, this task helps
us to achieve a list of “genes + variations”
(mapping) that are curated and validated, which in
the future will guarantee a diagnosis more reliable
and with higher quality.
3.3 Load
The loading process is done with the database
schema presented in the figure 3 (Martín M., 2011):
Table 1: View of Database (version 3).
Table Description
Structural
Genome
Version of DNA with which to work.
Chromosome
Elements that form the DNA sequence
chromosomes.
Chr_Elem
Stores each of the elements belonging to
a
chromosome.
Gene
Gene table (Table Chr_Elem specialization) is
defined as one of the most important
b
asic units
within the chromosome, and containing the
information necessary for the synthesis o
f
macromolecules, proteins usually with a specific
cellular function.
Sequence_Ng
Reference sequences of each of the genes foun
d
in the table stored Gene.
Transcript
Chromosome element that stores the resulting
transcript of the transcription process.
E
xon
Basic unit forming the mRNA transcript.
E
xon_Transcript
Associating table which serves to form part o
f
the transcript exons reference has an associate
d
gene.
P
rotein
Result of the translation of the mRNA for those
genes that code for protein.
Variations
Variation
Is the main table in this view and represents the
differences between different individuals.
P
recise
Variation specialization table representing the
variations detected with known position within
the chromosome in the DNA sequence.
P
recise_SeqNg
Is the variation that is associated with the
p
osition that such variation is compared to
a
reference sequence of a gene.
P
henotype
Phenotype is associated with the one or more
DNA variations.
Consideration of the Population
P
opulation
With this table we can associate the variations
found for certain diseases, and thus see the
impact of variations against other when we
crossed the population aspect.
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
286
The Table 1 shows different views to the
database that we use at the moment of our selective
load.
Figure 3: Database Schema: Variation + Structural View.
Structural view: shows the structure of the
genome of organisms.
Variations view: models the knowledge
related to differences in the DNA sequence to
different individuals.
And finally, it presents the consideration of
treatment of the populations for variations, and thus
the ability to generate an early diagnosis for any
disease.
3.4 Exploitation
The exploitation is the last stage of the SILE
methodology and their aim is the generation of
content from the point of view of customers or
stakeholders. The exploitation presents the results at
the end of the previous stages.
These contents include: text, images, etcetera.
And they must be clear and concise, so that
stakeholders can understand and manipulate all the
information effectively.
In our case, the exploitation process was
conducted with the design and implementation of a
Web Service, called “GenesLove.Me” (Reyes,
2013), that facilitates the acquisition of genetic tests
direct to consumers (GenesLove.Me, 2015).
4 IMPLEMENTATION OF SILE
The experiments will be conducted to verify the
application of SILE to a Genomic Information
Systems (GeIS), and thus achieving in this way:
efficient, reliable and agile systems.
4.1 Improved Quality and Time
Optimization
In case 1, we want to see if we can improve SILE
applying such circumstantial quality in generating
diagnoses, as well as achieving optimize load times.
We implement SILE in the “selective load”,
which helps us to improve efficiency to generate the
results (more accurate results). Because with this
type of load we focused only on the variations
associated with a disease and have been highly
proven through various studies. The other form is
with the "massive load" (Reyes, 2013) which taken
all genes and variations that are found for a disease,
without scientific merit because they are in
study/research, and that is why the percentages of
Table 2: INFO of Genes+SNPs and Populations Associated with Alcohol Sensitivity [Source: GenesCard].
Use of GeIS for Early Diagnosis of Alcohol Sensitivity
287
these diagnoses tend to be lower and require more
work. The data treatment with the selective load,
allows us to create clean and reliable databases
(repositories).
We must have processes that ensure information
filtering, otherwise, we are going to create databases
that are loaded with irrelevant data, and which do
not add value when we generated the diagnosis
(results). So, the selective load helps us to maintain
repositories with quality and optimal approaches.
4.2 Population
With this experiment we identify important trends
that can add value to the database. After applying
the “load process” with the help of selective-SILE in
the previous study, we found that combining this
with aspects related to the population, we take
another step forward as it would generate a result of
greater accuracy and especially that could secure an
early diagnosis for end-users.
Through many studies we have found that there
are genes which develop more in one population
than another, and this has major implications in the
results, because when we take into account and work
with general variations for all individuals, we
likewise generalize the diagnosis. Now, when we
use the specific variations, and we take the most
affected population in consideration, results can be
more precise.
5 RESULTS
5.1 Improved Quality and Time
Optimization
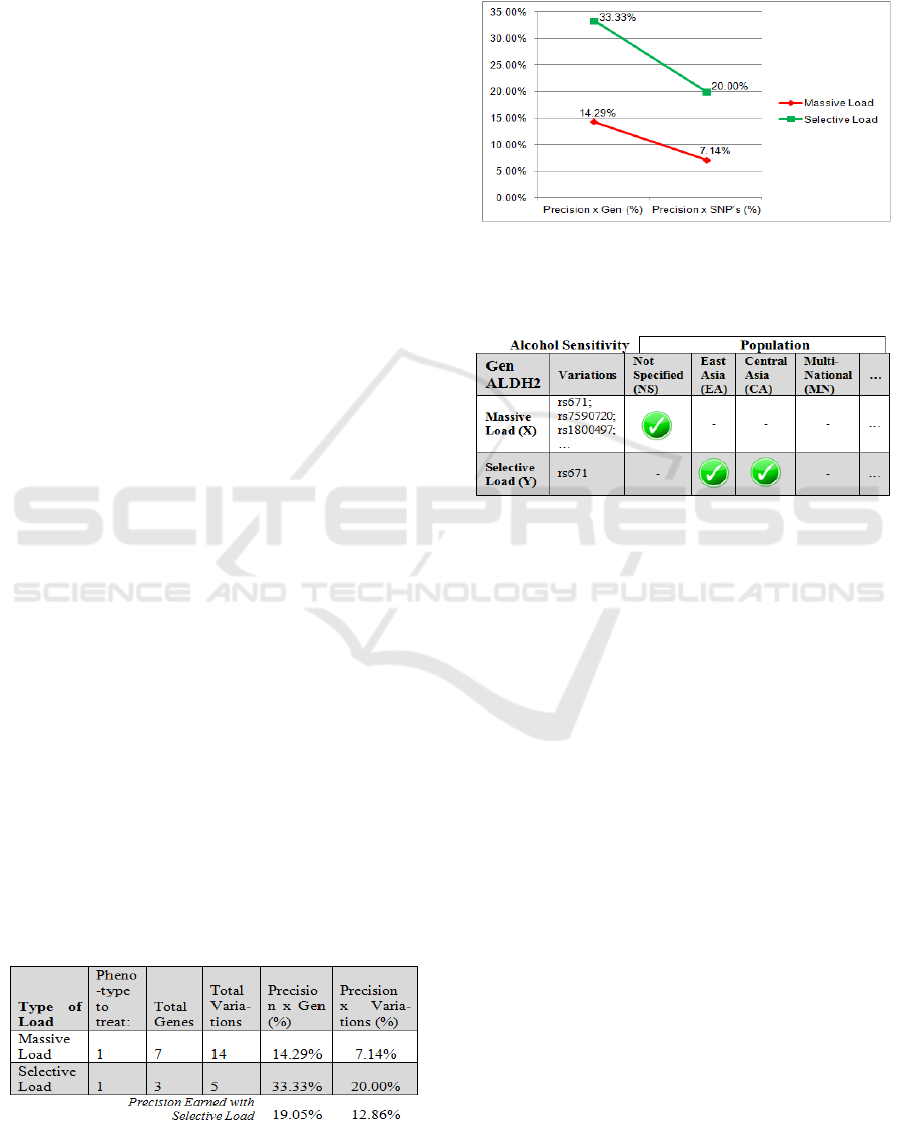
The use in our case of the selective load (Table 3),
increases genomic diagnostic quality when studying
a specific disease, and if we detected a small number
(curated) of gene + variations; we can obtain higher
precision percentages for the results (Figure 4).
Table 3: Precision Studies by Genes+Variations (Alcohol-
S).
5.2 Population
The study of population consisted of men and
women of full age, of different races and cultures.
Figure 4: Shows graph according to the type of load to
obtain accuracy level with respect to genes + variations.
Table 4: Genes and Variations by Population (Example).
When we use the massive load, it generates a
very general result, but turning to the selective load
it is possible to provide early diagnosis, simply
setting the initial observation of an ethnic group or
population of origin (Table 4).
6 CONCLUSIONS AND FUTURE
WORK
With the application of the SILE methodology we
obtained positive results, which as shown can be
improved circumstantially. Through our experiments
we have discovered key trends that help us add value
to our database.
We have concluded that we obtain an increase in
quality and optimization time using the selective
load, as we have the necessary data to generate
diagnoses, instead of having a database loaded with
all the information they provide, which is not
curated (filtered) and much of it without medical
validation.
We have also learned that by including
population information to previous studies, we can
deepen the diagnostics and take a step forward, since
there are variations of genes that affect some
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
288

populations more than others.
The implementation of this allows us to generate
and provide to the end-users an early diagnosis
about any disease (of genetic origin) with great
quality. The future work is oriented in the
implementation of the haplotypes for the human
genome database (HGDB). These are of great
interest because there are diseases that are diagnosed
by the particular combination of alleles for two or
more SNPs (Single-nucleotide polymorphism) that
are in the same chromosome. It is important in the
genetic diseases that are identified by the haplotype
variation and not as a single variation.
The development and growth in this area is
beneficial in the generation of diagnostics, and
particularly in the incorporation of
biopharmaceuticals for treatment and prevention to
the end-users. As the years pass, new variations are
considered to be associated with the alcohol
sensitivity, and it is only a matter of further research
and analysis of new samples that would allow the
medical community to give the approval for new
genes/variations.
ACKNOWLEDGEMENTS
The author thanks Ainoha Martín Mayordomo,
Mercedes Rossana Fernández Alcalá, David Roldán
Martínez and Edgars Groza for critically reading this
manuscript. We also thank the members of the
PROS Center Genome group for fruitful discussions.
In addition, it is also important to highlight that
this work has been supported by the Ministry of
Higher Education, Science and Technology
(MESCyT). Santo Domingo, Dominican Republic.
REFERENCES
Sherry, S. T., Ward, M. H., Kholodov, M., Baker, J., Phan,
L., Smigielski, E. M., & Sirotkin, K., 2001. dbSNP:
the NCBI database of genetic variation. Nucleic acids
research, 29(1), 308-311.
Viewing and using NCBI, 2015. NCBI National Center for
Biotechnology Information.
http://www.ncbi.nlm.nih.gov/
OMIM (Online Mendelian Inheritance in Man), reviewed
in NCBI, 2015. NCBI OMIM.
http://www.ncbi.nlm.nih.gov/omim.
Hamosh, A., Scott, A. F., Amberger, J. S., Bocchini, C.
A., & McKusick, V. A., 2005. Online Mendelian
Inheritance in Man (OMIM), a knowledgebase of
human genes and genetic disorders. Nucleic acids
research, 33(suppl 1), D514-D517.
Hubbard, T., Barker, D., Birney, E., Cameron, G., Chen,
Y., Clark, L., & Clamp, M., 2002. The Ensembl
genome database project. Nucleic acids research,
30(1), 38-41.
Escarabajal, M. D., 2003. Alteraciones genéticas
relacionadas con el alcoholismo. Rev Neurología, 37,
471-80.
Bierut, L. J., 2011. Genetic vulnerability and susceptibility
to substance dependence. Neuron, 69(4), 618-627.
Wang, J., Yuan, W., & Li, M. D., 2011. Genes and
pathways co-associated with the exposure to multiple
drugs of abuse, including alcohol,
amphetamine/methamphetamine, cocaine, marijuana,
morphine, and/or nicotine: a review of proteomics
analyses. Molecular neurobiology, 44(3), 269-286.
Baxevanis, A. D., & Ouellette, B. F., 2004.
Bioinformatics: a practical guide to the analysis of
genes and proteins (Vol. 43). John Wiley & Sons.
Villanueva, M. J., Guzmán, A. R., Valverde, F., & Levin,
A. M., 2012, May. Diagen: A model-based
bioinformatic tool for genetic analysis. In Research
Challenges in Information Science (RCIS), 2012 Sixth
International Conference on (pp. 1-2). IEEE.
Villanueva Del Pozo, M. J., 2011. Diagen: Modelado e
Implementación de un Framework para el Análisis
Personalizado del ADN. Tesis de Máster en Ing.
Software, Métodos Formales & Sistemas de
Información, Universitat Politècnica de València,
Valencia, España.
Dawyndt, P., Dedeurwaerdere, T., & Swings, J., 2006.
Exploring and exploiting microbiological commons:
contributions of bioinformatics and intellectual
property rights in sharing biological information.
Introduction to the special issue on the microbiological
commons. International Social Science Journal, 188,
249-258.
Reyes Román, J. F., 2013. Integración de Haplotipos al
Modelo Conceptual del Genoma Humano utilizando la
metodología SILE. Tesis de Máster en Ing. Software,
Métodos Formales & Sistemas de Información,
Universitat Politècnica de València, Valencia, España.
Goetz, T., 2007. 23andMe will decode your DNA for
$1,000: welcome to the age of genomics. Wired Mag,
(15).
GenesCard, 2013. GenesCard Main Page.
http://www.genecards.org/
Geneslove.me: Information about genetic tests offered.
Geneslove.me. http://geneslove.me/index.
Genotest Information & Services, (2015). Genotest.
http://www.trkgenetics.com/genotest.
Martín Mayordomo, A., 2011. Integración de Bases de
Datos Genómicas: Una Aproximación Basada en
Modelado Conceptual. Tesis de Máster en Ing.
Software, Métodos Formales & Sistemas de
Información, Universitat Politècnica de València,
Valencia, España.
Martin, A.; Celma, M., 2011. Integrating Human Genome
Variation Data: An Information System Approach,
International Workshop on Database and Expert
Systems Applications, DEXA, pp.65-69.
Use of GeIS for Early Diagnosis of Alcohol Sensitivity
289
