
Ontology based Description of Analytic Methods for Electrophysiology
Jan
ˇ
Stebet´ak
1
and Roman Moucek
2
1
Department of Computer Science and Engineering, University of West Bohemia, Univerzitni 8, Pilsen, Czech Republic
2
NTIS - New Technologies for the Information Society, University of West Bohemia, Univerzitni 8, Pilsen, Czech Republic
Keywords:
Neuroinformatics, Electroencephalography, Event-related Potentials, Analytic Methods, Metadata, Semantic
Web, Ontology.
Abstract:
The growing electrophysiology research leads to the collection of large amounts of experimental data and con-
sequently to the broader application, eventually development of analytic methods, algorithms, and workflows.
Then appropriate metadata definition and related data description is critical for long term storage and later
identification of experimental data. Although a detailed description of electrophysiology data has not become
a commonly used procedure so far, publicly available and well described data have started to appear in profes-
sional journals. The next reasonable step is to shift attention to the analysis of electrophysiology data. Since
the analysis of this kind of data is rather complex, identification and appropriate description of used methods,
algorithms and workflows would help reproducibility of the research in the field. This description would also
allow developing automatic or semi-automatic systems for data analysis or constructing complex workflows
in a more user friendly way. Based on these assumptions authors present a custom ontology for description
of analytic methods and workflows in electrophysiology that is proposed to be discussed within the scientific
community.
1 INTRODUCTION
Our research group at the Universityof West Bohemia
in Pilsen specializes in the research of human brain
activity. We use the methods and techniques of elec-
troencephalography (EEG) and event-related poten-
tials (ERP). As the electrophysiology research grows,
larger amounts of data are collected and more com-
plex methods and workflows are used for data analy-
sis. Typically, electrophysiology workflows used in
our laboratory include preprocessing methods (e.g.
filtering, baseline correction), signal processing meth-
ods (for example feature extraction methods, cluster-
ing and classification methods), and postprocessing
(usually statistical) methods. To make the applica-
tion of complex analytic methods and workflows re-
producible, a need for identification and sharing of
their appropriate descriptions arises. These descrip-
tions would also allow developing automatic or semi-
automatic systems for data analysis or constructing
complex workflows in a more user friendly way. Then
by defining proper metadata structures for analytic
methods and workflows and by using suitable tech-
nologies for machine-processing the workflow sys-
tems can ensure the following procedures:
• to check the syntactic compatibility of the used
methods (the output of the previous method is
used as an input to the following method)
• to check the semantic compatibility of the used
methods (connection of methods has sense in
terms of their semantic usage within the process-
ing chain and also semantic compatibility of trans-
ferred parameters)
• to suggest, which method is suitable to be put next
into the workflow
In this paper, we present a definition of the meta-
data structure that describes the methods used in ana-
lytic processing of electrophysiology data in our lab-
oratory. This structure thus helps to identify the se-
mantic compatibility of the methods while construct-
ing workflows. The rest of this paper is organized
as follows: Section II describes the analytic methods
that we use for processing of electrophysiology data.
It also briefly presents the Semantic web technolo-
gies and existing ontologies. Section III deals with
the metadata definition. The selection of a suitable
technologyis provided in Section IV and the proposed
ontology is presented in Section V. This section is fol-
lowed by conclusions and future work description.
420
Štebeták, J. and Moucek, R.
Ontology based Description of Analytic Methods for Electrophysiology.
DOI: 10.5220/0005814004200425
In Proceedings of the 9th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2016) - Volume 5: HEALTHINF, pages 420-425
ISBN: 978-989-758-170-0
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

2 STATE OF THE ART
This section brings an overview of existing ap-
proachesto description of analytic methods and work-
flows. Then it introduces the analytic methods suit-
able for electrophysiology research that we use for
data analysis in our laboratory. Then the Semantic
web technologies are briefly described.
2.1 Existing Approaches
The CARMEN Portal (Watson et al., 2007)
(Code Analysis, Repository & Modelling for e-
Neuroscience) developed by the British National
Node allows neuroscientists to save and share exper-
imental data and services. CARMEN provides stor-
age of services. There is a number of public services
available such as data filters, neural spike detection
and spike sorting methods. No formal semantics or
metadata structures are used for description of these
methods, they are described in a natural language.
The Galaxy project (Goecks et al., 2010);
(Blankenberg et al., 2010); (Giardine et al., 2005) is
an open source workflows engine. A registered user
is able to use methods and workflow tools provided
by this system. Galaxy is focused on genome analy-
sis; therefore, this system contains methods suitable
for genome analysis. The methods are well described
for the users with description of parameters and ex-
amples. It also includes OBI ontology (ontology for
biomedicalinvestigation)for formaldescription of se-
mantic restrictions.
The Wings is an open source workflows engine
(its source code is available at GitHub) and portal
based system downloadable and runnable on local
servers. The methods the engine works with are
described by ontologies (in the Resource Descrip-
tion Framework). This description includes defini-
tion of semantic constraints such as allowed values,
input/output cardinality, range, etc. The main disad-
vantage is a small community of developers and an-
noying bugs (e.g. difficulty with adding a new method
into the Wings system) that we found while trying to
deploy the system on a server.
This overview shows that the formal description
of methods in terms of semantic constraints (e.g. al-
lowed values) exists. However, the proper description
of input/output parameters allowing semantic com-
parison of methods is still not satisfactorily solved.
Therefore, we propose description of methods that al-
lows such comparison in this paper.
2.2 Analytic Methods
In our laboratory we widely use the following sig-
nal preprocessing and processing methods: averag-
ing, FIR filters, Matching Pursuit, Discrete and Con-
tinuous Wavelet transform, Fast Fourier transform,
Hilbert-Huang transform, and various neural net-
works. This section briefly describes the basic prin-
ciples of these algorithms since metadata definitions
that follow will be proposed just for these methods.
We admit that the methods described in this paper are
only a subset of a larger set of methods that can be
used in signal preprocessing and processing. This is
taken into account and the proposed design will allow
straightforward extension of metadata definitions.
Averaging (Sanei and Chambers, 2007) is a com-
mon method for highlighting ERP waveforms. Dur-
ing the averaging of the same kind of ERP waveforms,
the noise is reduced and the waveform is highlighted.
Since the background EEG has a higher amplitude
then ERP waveforms, the averaging technique high-
lights the waveforms and suppress the background
EEG (Vidal, 1977).
The Matching Pursuit method has been frequently
used for continuous EEG processing. It decomposes
any signal into a linear expansion of functions. At
each iteration, a waveform is chosen in order to best
match the significant structures of the signal. Typ-
ically, this part is approximated by a Gabor atom,
which has the highest scalar product with the origi-
nal signal, and then it is subtracted from the signal.
This process is repeated until the whole signal is ap-
proximated by Gabor atoms with an acceptable er-
ror (Vareka, 2012). For displaying results we imple-
mented the time-frequency transformation known as
Wigner-Ville transformation (Quian, 2002). The in-
put of this transformation is the set of chosen atoms.
Wavelet Transformation (WT) (Ciniburk et al.,
2010) is a suitable method for analyzing and process-
ing non-stationary signals such as EEG. WT has a
good time and frequency localization, which is nec-
essary for ERP detection. For EEG signal processing
it is possible to use Continuous Wavelet Transforma-
tion or Discrete Wavelet Transformation. For visual-
ization of wavelet results, the scalogram (Figure 1) is
used.
Figure 1: Input signal and its scalogram. (Rondik, 2012).
Ontology based Description of Analytic Methods for Electrophysiology
421

The Fourier transform converts waveform data in
the time domain into the frequency domain. Since
artifacts usually have higher amplitude and basic fre-
quency than a normal ERP component, this technique
is useful for detecting artifacts within the EEG or ERP
signal.
Independent Component Analysis (ICA) (Hyvari-
nen et al., 2001) is a method for blind signal sep-
aration and signal deconvolution. In the EEG/ERP
domain, ICA can be used for artifact removal, ERPs
detection, and, generally speaking, for detection and
separation of every signal which is independent on
EEG activity.
The Hilbert-Huang transform (HHT) was de-
signed to analyze nonlinear and non-stationary signal.
This also includes detection of ERP waveforms that is
described in (Ciniburk, 2011).
2.3 Semantic Web Technologies
The Semantic Web is a layered architecture. The
first layer is called Resource Description Frame-
work (RDF). RDF is a simple metadata representa-
tion framework using URIs to identify web-based re-
sources and a graph model for describing relation-
ships between resources. Web ontology language
(OWL) is a semantically richer language and provides
more complexconstraints on the types of resource and
their properties. OWL comes with a larger vocab-
ulary, greater machine interpretability and stronger
syntax than RDF.
There are substantial differences between clas-
sic object-oriented languages such as Java or C#
and Semantic Web technologies. The semantics of
classes and instances in RDF Schema is open-world
and description logics-based while object-oriented
type systems are closed-world and constraint-based
(A. Kalyanpur and Padget, 2002). The following
list brings main differences between OOP (Object
Oriented Programming) and Semantic web princi-
ples (Oren et al., 2007):
• class membership: in object-oriented languages,
an object is a member of exactly one class: its
membershi is fixed and is defined during the ob-
ject instantiation. In RDF Schema, a resource can
belong to multiple classes: its membership is not
fixed but defined by its rdf:type and the properties
that belong to the resource.
• class hierarchy: in object-oriented type systems,
classes can usually inherit from one superclass,
while in RDF Schema classes can inherit from
multiple superclasses.
• attribute vs. property: in the object-oriented
model, attributes are defined locally inside their
class, can be used only by instances of that class,
and generally have single-typed values. In con-
trast, RDF properties are stand-alone entities that
can be used by any resource of any class and that
can have values of different types.
• structural inheritance: in object-oriented pro-
gramming, objects inherit their attributes from
their parent classes. In RDF Schema, since prop-
erties do not belong to a class, they are not inher-
ited. Instead, property domains are propagated,
but given their specific meaning indicating the
class membership of resources using that prop-
erty, domains propagate into the upwards direc-
tion of the class hierarchy.
• object conformance: in most object-oriented lan-
guages, the structure of instances must exactly
follow the definition of their classes, whereas in
RDF Schema, a class definition is not exhaus-
tive and does not constrain the structure of its in-
stances: any RDF resource can use any property.
• flexibility: object-oriented systems usually do not
allow class definitions to evolve during runtime.
In contrast, RDF is designed for integration of het-
erogeneous data with varying structure from vary-
ing sources, where both schema and data evolve
during runtime.
The main advantage of Semantic Web technolo-
gies (e.g. RDF or OWL language) is the ability to
evolve during runtime. Since newly created or added
methods have to be well described, an extendable
metadata definition is necessary. Easy reusability of
classes and properties is also crucial, therefore the
Semantic Web concept and technologies were cho-
sen for description of the analytic methods described
above.
3 METADATA DEFINITION AND
ONTOLOGY DEVELOPMENT
Because there is no suitable description of the meth-
ods used in electrophysiology, we proposed their se-
mantic description by using a set of metadata. De-
scribing analytic methods at a more specific level for
workflow construction requires a detailed analysis of
the methods’ operations in terms of semantics of their
inputs and outputs.
The metadata identification originated from our
experience with data analysis, expertise of co-workers
from cooperating institutions, books describing prin-
ciples of EEG/ERP design and data recording (e.g.
(Steven, 2005)), and numerous scientific papers de-
HEALTHINF 2016 - 9th International Conference on Health Informatics
422
scribing processing of EEG/ERP data. We defined the
following metadata and their structure:
• Method - It describes a method including its name
and input/output types. It also includes definitions
of restrictions (e.g. name is a string).
• Input/Output- It describes input/outputtypes used
by analytic methods. It includes signal such as
EEG or ECG, and its restrictions (e.g. signal has
values of double), coefficients from the Wavelet
transform, or atom provided by the Matching Pur-
suit method (the atom is composed of scale, fre-
quency, modulus, phase, and position represented
as double values).
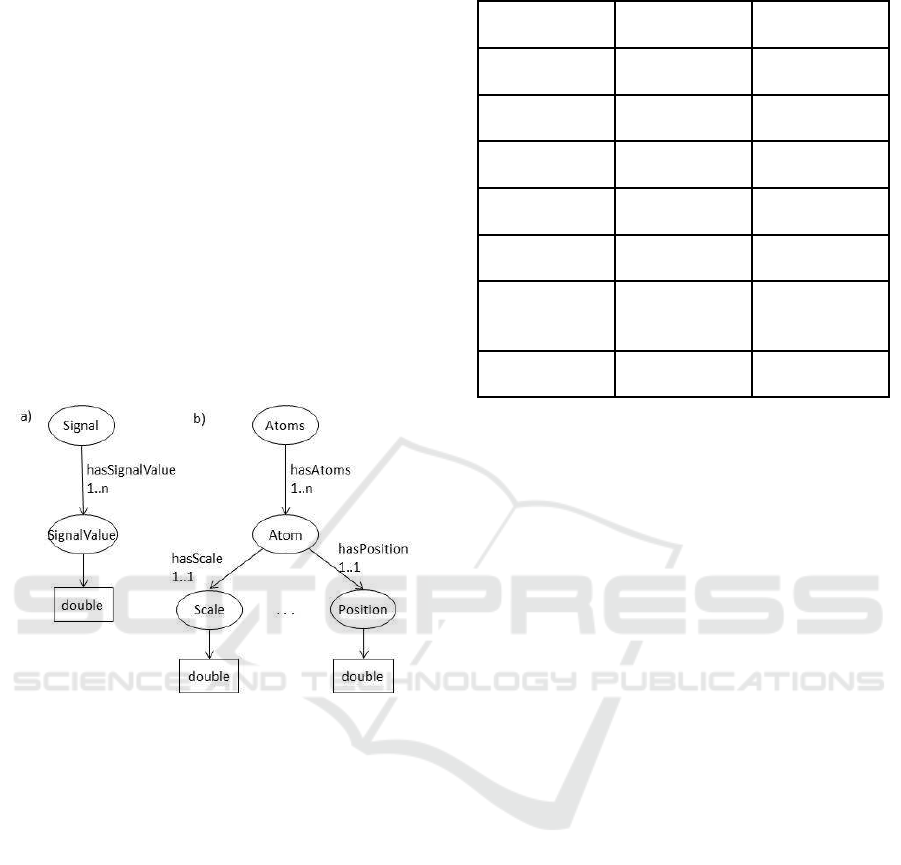
Figure 2 shows an example of the structure of in-
put/output types (Signal and Atom). The ovals repre-
sent elements and rectangles represents data types
Figure 2: a) Structure of Signal as an I/O type, b) Structure
of Atom.
Defining metadata we continued with the devel-
opment of the ontology for analytic methods intro-
duced in Section 2.2. Table 1 brings an overview
of these methods enriched with input/output param-
eter(s) types.
For the ontology development we used Protege,
which is a free, open-source ontology editor and
framework for building intelligent systems. The fol-
lowing terms were defined for the semantic group
method:
• Class Method and its named individuals (Detec-
tionOfEpochs, Averaging, CWT, DWT, FFT, Fas-
tICA, FIR, and MatchingPursuit)
• Class IOType representing general input/output
type (any class or individual)
We also defined properties hasInput and hasOut-
put. The class Method is a domain class for these
properties and the class IOType defines their range.
The class IOType has the following subclasses:
Table 1: Overview of methods and parameters.
Method name Input parame-
ter(s)
Output pa-
rameter(s)
Detection of
epochs
EEG signal Set of de-
tected epochs
Fourier
Transform
EEG/ERP
signal
Detected fre-
quencies
Wavelet
Transform
EEG/ERP
signal
Computed co-
efficients
Matching
Pursuit
EEG/ERP
signal
List of se-
lected atoms
ICA EEG/ERP
signal
ICA compo-
nents
Hilbert-
Huang Trans-
form
EEG/ERP
signal
Set of In-
trinsic Mode
Functions
Neural net-
work
Featured vec-
tor
Vector of
weights
• Class Signal (an example of this class is given
below) represents a set of values (signal ampli-
tudes) and individuals such as eegSignal, ecgSig-
nal, ...). Epoch, FilteredSignal, and Reconstruct-
edSignal are subclasses of this class. For these
classes we defined an object property hasSignal-
Value, which range is the class SignalValue lead-
ing to a data property named Value with the de-
fined type xsd:double.
• Class Coefficient represents a generic type for a
coefficient (for classes in this list that include co-
efficients as their object properties). The coef-
ficient value defined as object property is of the
double value.
• Classes CWTCoefficients and DWTCoefficients
are computed coefficients that represent the return
type of the methods Continuous Wavelet Trans-
form resp. Discrete Wavelet Transform.
• Class ComplexFrequency represents composi-
tion of RealFrequency and ImaginaryFrequency
classes. This type is typically used as an output
of time-frequency analytic methods such as Fast
Fourier Transform. It has two object properties:
hasRealValue and hasImaginaryValue. The prop-
erties range are RealValue class resp. Imaginary-
Value class.
• Class Atom represents the output type of the
Matching Pursuit method. It includes five coef-
ficient types (classes Frequency, Scale, Modulus,
Position, and Phase) and five corresponding ob-
ject properties (hasScale, hasPosition, etc.).
Below an RDF example containing the class Sig-
nal, the object property hasSignalValue, and the data
property Value is shown.
Ontology based Description of Analytic Methods for Electrophysiology
423

<owl:Class rdf:about=
"http://www.semanticweb.org/eegMethods#Signal">
<rdfs:subClassOf>
<owl:Restriction>
<owl:onProperty rdf:resource=
"http://www.semanticweb.org/
eegMethods#hasSignalValue"/>
<owl:onClass rdf:resource=
"http://www.semanticweb.org/
eegMethods#SignalValue"/>
<owl:minQualifiedCardinality
rdf:datatype="&xsd;
nonNegativeInteger">1
</owl:minQualifiedCardinality>
</owl:Restriction>
</rdfs:subClassOf>
</owl:Class>
<owl:ObjectProperty rdf:about=
"http://www.semanticweb.org/
eegMethods#hasSignalValue">
<rdfs:domain rdf:resource=
"http://www.semanticweb.org/
eegMethods#ICAComponent"/>
<rdfs:domain rdf:resource=
"http://www.semanticweb.org/
eegMethods#Signal"/>
<rdfs:range rdf:resource=
"http://www.semanticweb.org/
eegMethods#SignalValue"/>
</owl:ObjectProperty>
<owl:DatatypeProperty rdf:about=
"http://www.semanticweb.org/eegMethods#Value">
<rdfs:domain rdf:resource=
"http://www.semanticweb.org/
eegMethods#CoefficientValue"/>
<rdfs:domain rdf:resource=
"http://www.semanticweb.org/ontologies/
eegMethods#SignalValue"/>
<rdfs:range rdf:resource="&xsd;double"/>
</owl:DatatypeProperty>
Figure 3 shows a tree of defined classes provided by
the Protege.
The structure of the ontology including defined
terms, equivalent classes, subclasses, and used prop-
erties is designed for semantic comparison of in-
put/output parameters. It is suitable for looking for
the semantic similarity between input/output parame-
ters of the consecutive methods. This will ensure the
semantic compatibility of methods while putting them
into a workflow.
4 CONCLUSIONS
This paper presents a subset of methods suitable for
EEG/ERP signal preprocessing and processing that
are used in our laboratory. The basic principles of
these methods as well as their use for ERP waveforms
Figure 3: Tree of classes in Protege.
detection or artifacts removal are briefly described.
Since complex data analyses often require using mul-
tiple methods sequentially, it is crucial to find suit-
able methods and reasonable workflows to achievethe
goal. Therefore, we defined a set of metadata describ-
ing these analytic methods to help analyst to create
such a workflow correctly and with comparably lower
effort. Adding semantics to the methods in the form
of metadata thus allows analysts or better appropriate
software tools to check both syntactic and semantic
compatibility. Therefore we developed the ontology
that included a set of defined metadata. This brings
an ability to develop an automatic or semi-automatic
workflow system in future.
The presented ontology is also easily extendable.
It allows reusing existing classes and properties as
well as defining new classes and/or properties when
necessary.
4.1 Future Work
We will annotate the input/output parameters of the
methods used in our laboratory with the terms defined
in the ontology. It allows us to ensure the compatibil-
ity of methods while constructing workflows. We also
plan to develop a workflow suggestion system based
on the presented ontology that will help users to find
suitable methods while constructing workflows.
HEALTHINF 2016 - 9th International Conference on Health Informatics
424

ACKNOWLEDGEMENTS
The work was supported by the UWB grant SGS-
2013-039 Methods and Applications of Bio- and
Medical Informatics and by the European Regional
Development Fund (ERDF), Project ”NTIS - New
Technologies for Information Society”, European
Centre of Excellence, CZ.1.05/1.1.00/02.0090.
REFERENCES
A. Kalyanpur, D. Pastor, S. B. and Padget, J. (2002). Au-
tomatic mapping of owl ontologies into java. In Pro-
ceedings of the International Conference on Software
Engineering and Knowledge Engineering (SEKE).
Blankenberg, D., Kuster, G. V., Coraor, N., Ananda, G.,
Lazarus, R., Mangan, M., Nekrutenko, A., and Taylor,
J. (2010). Galaxy: A web-based genome analysis tool
for experimentalists. Current protocols in molecular
biology, pages 19–10.
Ciniburk, J. (2011). Hilbert-huang transform for erp detec-
tion. ph.d. thesis. Technical report, University of West
Bohemia, Department of Computer Science and Engi-
neering, Czech Republic.
Ciniburk, J., Moucek, R., Mautner, P., and Rondik, T.
(2010). Erp components detection using wavelet
transform and matching pursuit algorithm. In DCII,
Prague.
Giardine, B., Riemer, C., Hardison, R. C., Burhans, R., El-
nitski, L., Shah, P., Zhang, Y., Blankenberg, D., Al-
bert, I., Taylor, J., Miller, W. C., Kent, W. J., and
Nekrutenko, A. (2005). Galaxy: a platform for inter-
active large-scale genome analysis. Genome research,
15(10):1451–1455.
Goecks, J., Nekrutenko, A., Taylor, J., and Team, T. G.
(2010). Galaxy: a comprehensive approach for sup-
porting accessible, reproducible, and transparent com-
putational research in the life sciences. Genome Biol,
11(8):R86.
Hyvarinen, A., Karhunen, J., and Oja, E. (2001). Indepen-
dent component analysis. In Adaptive and Learning
Systems for Signal Processing, Prague.
Oren, E., Delbru, R., Gerke, S., Haller, A., and Decker, S.
(2007). Activerdf: object-oriented semantic web pro-
gramming. In WWW ’07: Proceedings of the 16th
international conference on World Wide Web, pages
817–824, New York, NY, USA.
Quian, S. (2002). Introduction to time-frequency and
wavelet transforms. Paris.
Rondik, T. (2012). Methods for detection of erp waveforms
in bci systems. state of the art and concept of ph.d.
thesis. Technical report, University of West Bohemia,
Department of Computer Science and Engineering.
Sanei, S. and Chambers, J. A. (2007). Eeg signal process-
ing. Chippenham (Wiltshire): Antony Rowe Ltd.
Steven, J. L. (2005). An Introduction to the Event-Related
Potential Technique (Cognitive Neuroscience). A
Bradford Book.
Vareka, L. (2012). Matching pursuit for p300-based brain
computer interfaces. Prague.
Vidal, J. J. (1977). Real-time detection of brain events in
eeg. In Proceedings of the IEEE, volume 65, pages
633–641.
Watson, P., Jackson, T., Pitsilis, G., Gibson, F., Austin, J.,
Fletcher, M., Liang, B., and Lord, P. (2007). The car-
men neuroscience server. In Proceedings of the UK
e-Science All hands Meeting, pages 135–141.
Ontology based Description of Analytic Methods for Electrophysiology
425
