
Sequence-based MicroRNA Clustering
Kübra Narcı
1
, Hasan Oğul
2
and Mahinur Akkaya
3
1
Medical Informatics Department, Informatics, Middle East Technical University, Ankara, Turkey
2
Department of Computer Engineering, Faculty of Engineering, Başkent University, Ankara, Turkey
3
Department of Chemistry, Faculty of Arts and Sciences, Middle East Technical University, Ankara, Turkey
Keywords: MicroRNA, Sequence Clustering, Clustering Algorithms, Pair-wise Sequence Comparison Sequence
Similarity.
Abstract: MicroRNAs (miRNAs) play important roles in post-transcriptional gene regulation. Altogether,
understanding integrative and co-operative activities in gene regulation is conjugated with identification of
miRNA families. In current applications, the identification of such groups of miRNAs is only investigated
by the projections of their expression patterns and so along with their functional relations. Considering the
fact that the miRNA regulation is mediated through its mature sequence by the recognition of the target
mRNA sequences in the RISC (RNA-induced silencing complex) binding regions, we argue here that
relevant miRNA groups can be obtained by de novo clustering them solely based on their sequence
information, by a sequence clustering approach. In this way, a new study can be guided by a set of
previously annotated miRNA groups without any preliminary experimentation or literature evidence. In this
report, we presents the results of a computational study that considers only mature miRNA sequences to
obtain relevant miRNA clusters using various machine learning methods employed with different sequence
representation schemes. Both statistical and biological evaluations encourages the use this approach in silico
assessment of functional miRNA groups.
1 INTRODUCTION
MiRNAs are small, 20-22 nucleotides in length, non-
coding RNA products of the corresponding MIR,
miRNA transcribing, genes. They regulate encoding
machinery involving in cleavage or translational
events by precise sequence complementation to the
target RNA sequence (Bartel 2004; Lagos-Quintana
et al. 2001). Due to its crucial function in the cell
identification, miRNA sequences has a great
importance since the earliest breakthrough
accumulated. The let-7 is one of the early identified
miRNA. The role of the miRNA is very important in
function; controlling differentiation in C. elegans.
The let-7 family members generally involve in the
same processes such as controlling developmental
timing (Abbott et al. 2005). Recently, it is found out
that the sequence of the let-7 family members are
also well conserved. Moreover, some of the MIR
genes found to be polycistronic transcribed into
miRNAs and located into the same chromosomal
positions; they are called as miRNA clusters. In
some of the miRNA clusters a recognizable
sequence similarity is also known (Altuvia et al.
2005). miRNAs targeted into a specific mRNA
region are greeted through biogenesis which is
commonly specific into the organism. At the end of
its biogenesis RNA-induced silencing complex
(RISC) is formed, and by RISC binding regions the
miRNA sequence is used as template to complement
the target mRNA sequence. The consequence is
either miRNA cleavage by degradation or
translational repression by blocking the mRNA
being translated. Conversely, there would be a
positive result like sponsoring transcription or
translation and stabilization of transcription (Asgari
2011). In the complementation there are some key
regions important for target determination. Second to
eight nucleotides of the pre-miRNA sequence called
as seed region are known as key nucleotides (Bartel
2013). miRNA binding sites in target mRNA region
is generally in 3 ‘UTR region, occasionally in 5
‘UTR region of the gene in animals. The percentage
of the complementarities changes by, and depends
on type of the organisms (Pratt and MacRae 2009).
Through the evolutionary time, as the cell getting
change, miRNA to target relation diverse. From this
Narcı, K., O
˘
gul, H. and Akkaya, M.
Sequence-based MicroRNA Clustering.
DOI: 10.5220/0005552901070116
In Proceedings of the 9th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2016) - Volume 3: BIOINFORMATICS, pages 107-116
ISBN: 978-989-758-170-0
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
107

context, studies to observe sequence fingerprints in
miRNA families has been started (Hertel et al. 2012;
Shi et al. 2012) . The investigations on let-7 family
bring the consideration up that there are some
conserved patterns alongside with base composition
similarities between miRNA sequences (Hertel et al.
2012; Newman et al. 2008). Furthermore, it is found
that there is evolutionary importance of the
mismatches between miRNA and its target as well.
The mismatches are tolerating the easier release
from the RISC complexes when perfectly located
into its target (Bartel and Bartel 2003). All these
findings in the end support that there is a great
concurrence between the miRNA and target
sequence even evolutionary favoring the
mismatches. Hence, what is the level of these
similarity between existing miRNA groups?
It is well known that a number of miRNA
element may role in multiple functions, like
proliferation, cell death and differentiation,
immunity and fat metabolism by various pattern of
expression (He and Hannon 2004). Therefore, the
network including miRNA and its targets is highly
complex housing several genes. The analysis of
these relation may be compromised through
advanced tools like miRWalk2.0 (Dweep and Gretz
2015). The latest studies have shown that miRNAs
usually operate in a co-operative manner to perform
their activities (Antonov et al. 2009). This suggests
that some miRNAs can form context-specific
modules, i.e., cluster of entities, while regulating
gene expression. Since the elucidation of gene
regulatory networks comprising all actors is one of
the ultimate goals of systems biology, which
miRNAs are functionally similar in a certain context
is high-potential knowledge for the researchers and
clinicians working in this domain (Ölçer and Oğul
2013).
Recently, as the importance of miRNA directed
gene regulation become clear, computational
miRNA prediction tools was an active research area
(Zhao et al. 2010; Lai et al. 2003). Following the
advance, many predicted miRNAs sourced to be
characterized into function. Here in this study, in the
light of the current miRNA literature we used the
sequence clustering approach to group mature
miRNAs in order identification of miRNA families
acting in the same metabolic events. In
bioinformatics, the attempt of grouping the
biological sequences is not novel. USEARCH and
UBLAST (Edgar 2010)are two algorithms developed
recently in that concept operating on nucleic acid
sequences, and there are TribeMCL (Enright et al.
2002) and OrthoMCL (Li et al. 2003) operating on
amino acid sequences. Sequential simulation of each
miRNAs in like the mentioned studies presented by
numeric kernels constructed though dynamic
programming pair-wise sequence alignment
algorithms; Smith-Waterman (Smith and Waterman
1981) and Needleman-Wunsch (Needleman and
Wunsch 1970) or by calculating their k-mer
distributions. Unsupervised clustering approaches
then applied into these sequence simulations by
using the similarity features. The performance of the
clusters is statistically analyzed by using Dunn Index
(Dunn 1973) calculation and the functionality of the
pipeline is tested with a well-known human miRNA
dataset of Tool for Annotations of miRNA (TAM)
(Lu et al. 2010). The tool also used to test the
groups, annotate them into functional categories and
thus calculate the enrichment of the miRNA groups
with any purposeful similarities. In conclusion, the
workflow here represents the method to explore
sequentially similar miRNAs and their relevance in
groups.
2 MATERIALS AND METHOD
2.1 Clustering
The task is to assign each miRNA into one of
previously unlabeled classes so that a set of non-
overlapping miRNA groups, which are desired to
imply a useful relevance, can be obtained. This can
be achieved through an unsupervised machine
learning technique called as clustering (Flynn 1999;
Sisodia 2012). As having a large diversity of
clustering algorithms in machine learning society,
we consider here four distinct methods which are
selected based on their common use in
bioinformatics studies; k-means (Macqueen 1967),
CLAG (Dib and Carbone 2012), and SOTA (Dopazo
et al. 1997) and MCL (Enright et al. 2002), which
are briefly introduced as follows.
K-means (Macqueen 1967) is the classical yet
the most common in use method of partitional
clustering. By the technique, the dataset is divided
into k non-overlapping groups by means of
minimizing the sum of squares of distances between
data points and the corresponding cluster centroids.
The logic of the method depends on the iterations of
these steps; (1) determination of the centroid
coordinate, (2) evaluating the distance of each object
to the centroids and, finally (3) grouping into the
objects based on minimum distance (Macqueen
1967). Prior to these steps however, k must be
specified. Actually, if the dataset is unknown and
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
108

analyzer doesn’t know how many grouping will be
done, optimization of k becomes one of the
weaknesses of this method. Moreover, if the
numbers of data are not high enough, initial
groupings will determine the cluster contents
significantly. Therefore, with different centroids,
different classifications are possible and the
evaluation of validity of these clusters becomes
substantial (Rawlins et al. 2012). To overcome
different partition problem in our analysis, re-run
results are generated for the same input of cluster.
After memberships of the clusters defined, each trial
was compared to each other in order to detect most
stable groups. Therefore, some extension is made
through the clusters, and less stable groups divided if
their membership is not convincing enough for other
trials or if the member is unstable for an affiliation it
is eluted from the set as suggested by (Jain 2010).
CLAG (CLuster Aggregation)(Dib and Carbone
2012) method is specially established for large non-
uniform biological datasets. It is an unsupervised
non-hierarchical method aiming to zoom in only
compressed regions in the uneven datasets by given
parameters. The algorithm iterates for suitable
aggregations on the dense regions. Therefore, the
algorithm does not group whole data; instead, only
finds best similarities in the particular correlation
metrics. One of the benefits of the algorithm is that
the cluster number is not specified by the user.
Furthermore, since the algorithm does not samples
the data with initial centroids, it does not suffer from
the problems of k-means, like yielding different
clusters for repeat runs and dealing with dense-
shaped data points.
SOTA (Self-Organizing Three Algorithm)
(Herrero et al. 2001) is a hierarchical clustering
method unusually using neural network (Self-
Organizing Map- SOM) centered on a distance
function well fit to the nature of the data. Neural
network propagates to fit the topology of the set into
a binary tree. The algorithm aims to integrate
advantages of both methods hierarchical clustering
and SOM without suffering from their problems.
SOTA is a divisive method, clusters form from a
growing neural network, with respect to
agglomerative approach of hierarchical algorithms.
This feature of SOTA has led to stop at any desired
level of hierarchy until cluster numbers reach to
equality with data points, and so, arrangement of the
homogeneity of the clusters is arrived. Prior to the
analysis, the algorithm evaluates the distances
between the elements and chooses two main nodes.
The following divisions calculated up to
homogeneity of these nodes are absolute not change.
This makes the centroids of the data fixed; re-runs of
the data do not change the position of the centroids
and thus, with respect to k-means algorithm, cluster
members remains fixed (Dopazo et al. 1997; Herrero
et al. 2001).SOTA method is proven to cluster large
gene expression patterns like microarray analysis
results. The method is efficient to be able to isolate
the real clusters from the noise of the data (Herrero
et al. 2003).
MCL(Markov Clustering Algorithm) (Enright et
al. 2002) is a graph clustering method developed by
Stijin Von Dongen at 2000. This algorithm has been
widely used in bioinformatics to find functional
relations in protein datasets. Such as OrthoMCL (Li
et al. 2003) and TribeMCL (Enright et al. 2002) use
MCL algorithm applied into all-to-all BLAST results
of protein sequences. MCL algorithm uses a
weighted symmetry matrix which shows the pair-
wise distances between the items in the dataset. The
pair-wise weights are turned into transition
probabilities with normalizations. The algorithm
makes random walks using probability matrix to find
inter connected elements namely the clusters. In
general, the algorithm has two steps; normalization
and inflammation. Normalization step is responsible
for calculating probabilities of each connection for
each node in the graph. After each normalization
step, to overweight current strength connections and
on the contrary underweight the weak ones the
square root of the matrix is taken names as
inflammation. The inflammation value can be
arranged by the behavior and the structure of the
dataset. It can be increased to find more strength
connected clusters and to observe bi-connected
groupings, and it can also be decreased to find
naturally big connections or to present well
separated groups. These two steps, normalization
and inflammation, iterate on the graph until the
convergence is fixed (Enright et al. 2002; Li et al.
2003). The algorithm is very gainful on classical
vector based cluster algorithms when the distance
metric is considered as important between objects.
The method instantly found the cluster number
unlike the classical methods. Unlike k-means and
SOTA cluster number is not provided by the end
user.
2.2 Sequence Representation
In the study, two approaches are used to represent a
miRNA sequence in a machine learning framework.
In the first method, a sequence is composed a set of
elements, each of which denotes similarity of current
miRNA sequence with any other miRNA sequence
Sequence-based MicroRNA Clustering
109

in the repository. To quantize this resemblance, a
distance measure can be scored by pair-wise
alignment algorithms. To test different
methodologies, Smith-Waterman algorithm (Smith
and Waterman 1981) as local and Needleman-
Wunsch algorithm (Needleman and Wunsch 1970)
as global alignment are both applied. All sequences
in the list are aligned to each other in a pair-wise
manner, and their alignment scores are stored into a
symmetric all-to-all matrix. In the matrix, the nodes
demonstrate score vectors with respect to edges are
miRNA sequences (Similarity Matrix). To generate a
matrix showing distance measures (Distance Matrix)
, however, the scores for a miRNA sequence aligned
to other miRNAs is subtracted from the score
produced from the self-alignment of that miRNA,
basically that is the maximum score a miRNA
sequence can produce. When Needleman-Wunsch
algorithm is applied, negative scores are also
possible with respect to Smith-Waterman that creates
only positive scores. Therefore, the similarity and
dissimilarity (distance) matrices should not be
thought as real representative graphical distances,
instead, they are the metric values showing how two
pairings are alike or distant. Scoring schemes for the
both algorithms are the same, scores are calculated
according to Gap=-1, Mismatch=-1, and Match=+1
values.
The second method to deduce information from
sequence is to count k-mers. It aims to produce a
sequence model defined on distribution of k-mers,
namely all probable k length substrings. We chose k
as 3 for a 3-mer representation. On a defined RNA
alphabet (A, G, U, C), when k equals to 3, there is
4k, 64 distinct count values. The presence of 3
length substrings (like AGU, CAA, GAU…) can be
controlled and their presence indications can be
stated as 1 or unlikely situation can be 0.
Consequently, number of miRNAs versus 4k
dimension matrix is filled by 1 and 0. By this
method, sequence information becomes independent
from nucleotide triplet order, and the sequences are
not affected from each other (Oğul and Mumcuoğlu
2007).
2.3 Data
To assess the functional relevance of miRNA
clusters obtained through computational models, we
use a set of human miRNAs with experimentally
validated functional annotations. Current TAM
miRNA catalogue (Lu et al. 2010) for this purpose
provided miRNA sets for 413 distinct human
miRNAs. The miRNA groups in TAM are specified
in 5 distinct categories; family, function, tissue,
disease and cluster (genomic loci). A miRNA may
reside in more than one group provided that each
group is specified in a different category. In this
way, a set of overlapping miRNA groups can be
retrieved in varying annotation schemes. Family and
cluster specifications are based on miRBase
(Kozomara and Griffiths-Jones 2011) classes.
Human MiRNA Disease Database (HMDD) (Lu et
al. 2008) is used for disease specific associations,
and function and specific tissue relations is collected
from literature. In current version, TAM database
contains 238 miRNA sets in total. In TAM
repository, the names are not specialized with their
3’ or 5’ overhangs. Therefore, miRNA names are
matched with their corresponding sequences in
miRBase tool. When both overhangs are stored, final
dataset comprises 666 miRNA sequences in total.
2.4 Biological Validation of the
Clusters
For an agreed set of miRNAs, TAM tool estimates a
significance (p value) for each of its categories, and
this value describes the enrichment in the set. The
enrichment value is the function of TAM describes
how these miRNAs related depending on literature
reviews. P value is calculated in a correspondence
with the size of the given set of miRNA and size of
the dataset. Therefore, percentage of how many
given miRNAs are in a consistent cluster and its
significance are outputs of the tool (Lu et al.
2010).In our analysis, p-value (>0.005) and
percentage coverage (>0.2) are used to assess the
level of enrichments, only if there is two miRNA
found to be related. Each cluster for all clustering
method we used is tested by TAM, enriched clusters
are counted, and percentage of them calculated.
Therefore, the overall enrichment score is the
percentage of successfully enriched clusters per
given the total groupings
2.5 Statistical Validation of the
Clusters
Dunn Index (DI) calculation is used to get the ratio
of the smallest distance between the observations in
the different clusters to the largest distance of the
observations in the same cluster (Dunn 1973). DI
metric aims to signify how compact and well
separated the clusters is. The value of DI is 0 when
all of the objects are in the same cluster and infinite
when all the objects present for a cluster. To get a
better result, DI needs to be maximized. The distance
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
110
metric in DI can be classical Euclidian or Manhattan
distance. The method is used in order to understand
if the data is well separated prior to selection of
clustering parameters. Only clustered groups are
used in the study.
3 RESULTS AND DISCUSSION
3.1 Data Coverage and Cluster
Numbers
The first clue to be provided in order to understand
the compactness of the cluster is to determine cluster
numbers (number of grouping) and data coverage
(percentage of clustered miRNAs) (Table 1). As
CLAG algorithm tends to devise most strength
condense regions, it has the smallest data coverage
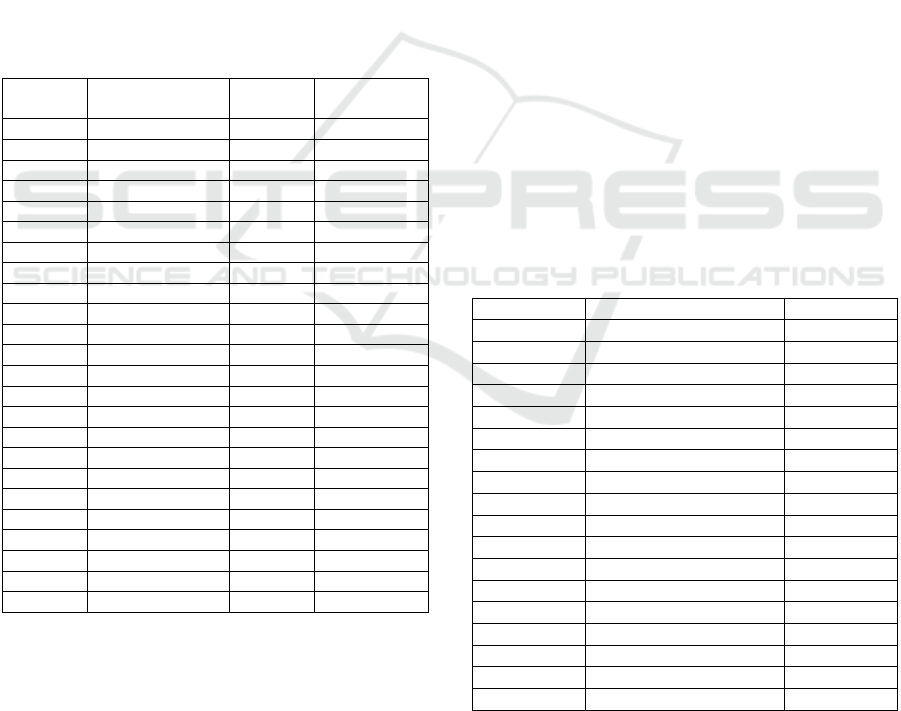
Table 1: Cluster numbers and Data Coverage’s (%) of the
groupings by different methods.
Matrix* Cluster
Number
Data
Coverage
K-means K-mer 47 99.85
NW-Similarity 46 85.44
NW-Distance 46 82.73
SW-Similarity 38 98.95
SW-Distance 37 96.55
Random Matrix 47 100.00
CLAG K-mer 29 9.16
NW-Similarity 30 10.96
NW-Distance 31 11.26
SW-Similarity 50 18.62
SW-Distance 24 8.56
Random Matrix 104 97.60
SOTA K-mer 30 100.00
NW-Similarity 30 100.00
NW-Distance 30 100.00
SW-Similarity 30 100.00
SW-Distance 30 100.00
Random Matrix 30 100.00
MCL A 15 86.04
B 18 73.12
C 17 63.81
D 56 75.96
E 46 58.41
F 46 52.70
*A, B and C are the 2
nd
; D, E, and F are the 4
th
power of
the original Smith Waterman applied MCL matrix.
4,5,6,2,3, and 4 inflation values are applied into
respectively A, B, C, D, E and F.
among the other stated methods. Thus, cluster
numbers and data coverage are very small (9% to
18%). Because of the same reason also, CLAG
operates different on Random matrix than the real
matrices. Random assignment of numbers generates
a scattered and district regions in the matrix which
CLAG cannot directly cluster the data. Which is
opposite of SOTA algorithm, it clusters the whole
dataset and set up of initial cluster number is through
manual. In a structural manner, classical K-means
algorithm also clusters the whole dataset. However,
monic clusters are also generated as district objects,
re-runs are required to remove the most district
elements. After several arrangements by DI
calculation, cluster number is optimally found as 43.
Random matrix results with 47 clusters and 100%
coverage.
MCL algorithm has a different methodology than
other algorithms since it is a graphical clustering
method. Data coverage is the value of inflammation
value. At least 15 number of clusters with 86% is
found for the matrix powered by 2 and inflamed by
4, and the most 56 number of clusters with 73% is
found for the matrix powered by four and inflamed
by 2 .
The representation of miRNA sequence was
differentiated in the cause of whether how
simulation important. Diverse matrices (Similarity,
Distance, and K-mer) implicated variation by cluster
numbers and data coverage, as like for also Simith-
Waterman and Needleman-Wunsch algorithms.
3.2 Statistical Validation
Table 2: Dunn Indexes evaluations for clusters.
Matrix Dunn Index
K-means K-mer 0.3511
NW-Similarity 0.2641
NW-Distance 0.2297
SW-Similarity 0.4539
SW-Distance 0.3507
Random Matrix 0.7920
CLAG K-mer 0.7454
NW-Similarity 0.6257
NW-Distance 0.4498
SW-Similarity 0.4867
SW-Distance 0.4789
Random Matrix 0.8311
SOTA K-mer 0.2970
NW-Similarity 0.2430
NW-Distance 0.2043
SW-Similarity 0.2766
SW-Distance 0.2369
Random Matrix 0.8396
Besides to data coverage and cluster numbers, an
arithmetic approach needed to calculate the strength
of the clusters. For this purpose, Dunn Index (DI)
values are evaluated (Table 2). Rather than other
Sequence-based MicroRNA Clustering
111

methods, DI is not considered with MCL algorithm
since it is a graph clustering method, similarity or
dissimilarity metric between objects can not indicate
the real distance values. The observation suggesting
that DIs overvalued for Random matrices can appear
as they are well grouped than real matrices. The
reason behind that is Random matrix is filled
unsystematically and so very homogeny that
contains no noise.
CLAG only objects into the condense regions,
finds small number of clusters, removing nearly 90%
of the data. Therefore, clusters are compact, as a
result DI values are better according other methods.
Among the two methods, K-means algorithm can
produce better clusters than SOTA for some
methods. SOTA DIs are low since SOTA tends to
cluster whole data unlike altered K-means algorithm.
Therefore, SOTA parcels the data without rendering
out the separated objects.
3.3 Biological Validation
The next step in the cause of asserting biological
function into miRNA clusters, the next step carried
through this study is categorization of the clusters by
TAM tool. All five categories of TAM tool is shown in
table 3; Clusters, Function, Family, HMDD, Tissue,
and ALL (as altogether), with respect to enrichment
results of clusters as percentages.
In order to significate a cut-off value, given the set
of miRNA random samples are taken and analyzed
through the same way by TAM tool (10, 30, and 150
grouping). Depending on sampling values, miRNA
enrichments change expressively. 10 clusters, for
example, tend to show increase in enrichment, whilst
150 clusters nearly does not give any enrichment
results. Actually, it is about the size of the cluster, as
size of a cluster increase, it is more likely it gets hit
from TAM tool. As the samples are only taken by
change, we can confirm the meaning of the grouping
generated by sequence similarity. Yet, our cluster
analyses in this study mostly run by nearly 30 number
Table 3: Enrichment results of clusters calculated by TAM tool.
Matrix Clusters Function Family HMDD Tissue All*
K-means K-mer 44.68 12.76 72.34 44.68 10.63 80.85
NW-Similarity 57.78 22.22 82.22 40.00 11.11 88.89
NW-Distance 52.17 19.56 76.09 45.65 6.52 84.78
SW-Similarity 63.16 23.68 78.94 55.26 15.79 92.11
SW-Distance 56.76 32.43 70.27 40.54 10.81 81.08
Random Matrix 8.51 6.38 8.51 14.89 4.25 34.04
CLAG K-mer 24.13 13.79 75.86 27.59 13.79 75.86
NW-Similarity 30.00 16.67 80.00 36.67 13.33 80.00
NW-Distance 22.58 16.13 77.42 41.94 9.68 77.42
SW-Similarity 20.00 14.00 70.00 24.00 10.00 70.00
SW-Distance 16.67 20.83 79.17 45.83 4.17 79.17
Random matrix 4.81 2.88 4.81 8.63 0 16.35
MCL A 60.00 26.67 86.66 33.33 13.33 86.66
B 38.89 22.22 66.67 33.33 16.67 72.22
C 41.17 23.53 58.82 29.41 17.65 70.59
D 32.14 14.29 51.79 25.00 7.14 66.07
E 39.13 13.04 47.03 21.74 8.70 60.87
F 39.13 15.22 47.83 21.74 10.87 60.87
SOTA K-mer 50.00 33.33 63.33 36.67 10.00 80.00
NW-Similarity 40.00 23.33 73.33 26.67 6.67 80.00
NW-Distance 36.67 13.33 70.00 50.00 6.67 83.33
SW-Similarity 50.00 30.00 70.00 43.33 10.00 83.33
SW-Distance 43.33 23.33 60.00 50.00 10.00 73.33
Random Matrix 10.00 10.00 10.00 20.00 3.33 43.33
Random Clusters 10 groups 10.00 13.33 6.67 33.33 0 46.67
30 groups 14.44 10.00 11.11 22.22 0 40.00
150 groups 3.16 4.28 2.48 9.93 0.68 16.7
* Five categories of TAM tool is shown; clusters, function, family, HMDD, and tissue. All represents the percentage result
annotated by any of the categories at least one time. The results are given as percentage. 30 groups chosen as cut-off value
bonded.
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
112

of clusters. Consequently, 40% percentage is chosen as
effective cut-off value. Furthermore, before, randomly
filled matrices were also generated to determine the
cluster sizes, they are also used as control metric
through analysis by TAM. For ALL case, Random
matrix enrichments are also found lower than 40%
percentage.
Two different similarity detection approaches, k-
mer counting and pair-wise sequence comparison, were
analyzed with TAM tool. Results illustrate that even
though, for k-mer method SOTA and k-means
algorithms are not effective as CLAG and MLC, the
general view to the outputs of TAM tool signifies that
there is no considerable change by modification in
similarity methods, at least 70% of clusters display
enrichment which is considerably exceeds the cut-off
value 40
%
. The finding proves that all of the similarity
representation methods able to show a momentous
enrichment over cut-off values (14%, 10%, 11%, 22%,
0, and 40% sequentially as Table 3).
Besides, no matter which method in which way is
used we see weighted enrichments for all 5 categories.
Also, between the categories of TAM, it is found out
that the most enrich one is family (60% to 86% with
11% cut off). This result proves the hypothesis that
among the same miRNA families there is a
considerable sequence similarity. For cluster category,
the enrichment is also noteworthy. In literature,
between the same clusters like let-7 family sequence
similarity is stated (Hertel et al. 2012; Newman et al.
2008). As the results estimate, we prove that there are
significant sequence similarities between some miRNA
clusters. However, in conclusion we see that miRNA
clusters itself are not correspond to a major sequence
similarity, unless they are not originated from the same
hairpin. Cluster enrichments are respectively smaller
since clusters of miRNAs are found by expression
analysis and proximity in location also. Nevertheless,
the information for Cluster category is not enough for
encompassing study, more literature views needed.
Furthermore, unavoidably tissue analysis of TAM was
not complete and so the information was not adequate
for a meaningful analysis, which is observable through
the results. Thus, to increase the quality of the
biological evaluation the collected information for
miRNA relations need to be large enough.
MCL method is applied only by Smith-Waterman
distance matrix. Various optimizations are needed to
made TAM enrichment analysis. Outputs show that
when a prior data inflation is increased, more clusters
are found but less functional annotation is possible
through TAM. Best functional annotation for Clusters
category (60%) is found by MCL algorithm with the
matrix powered by 2 and inflated with 4. This matrix
was the also less covered dataset, probably only found
the best relations in the dataset. MCL algorithm with
respect to other algorithms uses graph theory for
grouping indeed able to generate well group of
miRNAs separated from noise with 86% of enrichment
in function.
Data coverage found to be also related with
enrichment analysis. CLAG analysis represents that as
data coverage decrease, more similarity can be found in
miRNA sequences. Since only pair-wise similarities are
detected at least 75 % of the clusters enriched in all
categories and in families. K-means algorithm with
respect to CLAG tends to cover whole data and show
more enrichment. At least 80 % of the clusters enriched
in all categories. SOTA, like k-means, also show at
least 80% enrichment, but in cluster category, K-means
better than SOTA and any other cluster algorithms.
K-means and SOTA algorithms are able to cluster
whole data, with significant DI values. By using the
classical K-means algorithm, in fact, it is possible to
generate clusters 92% of them enriched in at least one
of TAM categories, and also 82% of them significantly
enriched in family category. However CLAG
algorithm only projects into condense regions of the
data, and found small major shrink clusters visualized
by low data coverage with high DI value. Yet, it is
proven that, these small clusters are well enriched in
function (80 % in ALL). Therefore, a pipeline can be
constructed as using CLAG a prior to cluster analysis to
shape the centroids of the data.
3.4 MicroRNA-target Relations
Here, the method describes how the miRNA sequences
can be clustered by using alone its sequence patterns.
Yet, as mentioned in methodology part, a miRNA can
regulate various mechanisms and processes in the cell
(Antonov et al. 2009). Recently, there are researches
focusing on specific miRNA to disease relations(Satoh
2012; Jacobsen et al. 2013), these are the touchstones of
broad searches on miRNA regulated gene networks
(Gennarino et al. 2012) . These studies suggest that
miRNA target gene ontology needs to be investigated
in a well-shaped network design. In our study,
nevertheless, as the complexity of the network for
many miRNA to many target relation make the
clustering of the targets very hard without designing a
new algorithm, we could not cover clustering of
miRNA target genes.
4 CONCLUSIONS
In search of finding miRNA groups with predicted
Sequence-based MicroRNA Clustering
113

functions regulating their target genes in turn pathways
such as development, immunity, environmental
responses and many more, the expression levels of
miRNAs are commonly preferred approach, since
experimental analyses are considered most reliable and
promising. However, they are indeed costly and time
consuming. Therefore, there is an urgent need in
generating computational tools for cluster analyses to
determine relevant miRNAs groups. Toward this end,
this study is focused on developing a novel approach
using the data available in databanks of human genome
with experimentally determined mature miRNA
sequences. Given a list of mature miRNA sequences,
sequence content translated into a metric system and
clustered by available clustering algorithms.
In this study, we provided a workflow for clustering
miRNA sequences independent from their expression
profiles using a sequence clustering approach by means
of existing machine learning algorithms, K-means,
CLAG, SOTA and MCL. Given a list of mature
miRNA sequences, similarity relations were detected
by two approaches; k-length substring counting and
pair-wise sequence alignment algorithms. To detect
pair-wise similarities between two sequences Smith-
Waterman and Needleman-Wunsch algorithms were
used. As a result, three different sequence
representation methodologies were utilized to detect
sequence similarities. Pair-wise sequence algorithms
were used to construct a matrix filled by scores of
descriptive scores. An all-to-all approach is used and all
sequences in the list compared to each other. Thus, the
filled matrix becomes the representations of distances
between all miRNAs, and it is used as input of cluster
algorithms. The other approach was k-mer counting,
independent from the order which is a priority in pair-
wise alignment algorithms. It is also a novel approach
for representation of a sequence as input of clustering
algorithms.
Preexisting clustering methods are used in the
contexts of the study in order to provide a comparison
between different methodologies which are appropriate
for different type of metrics. The methods used in this
study have been not previously applied into a miRNA
sequence metric matrices. In that perspective too, this
study has also an innovative outcome. Only, MCL
algorithm which is a graphical clustering method
indeed was originated to cluster protein sequence score
metrics, which is very useful for sequences represented
as distance values. From different perspective used in
this study, hierarchical clustering on nucleic sequences
is possible through multiple sequence alignment (MSA)
(Corpet 1988). Because of the fact that MSA methods
directly operate on sequences, but not on a metric in
matrix, in this study we did not used this standpoint.
Furthermore, within the supervision of the sequence
similarity information behind some clusters studies
before (Hertel et al. 2012; Abbott et al. 2005; Newman
et al. 2008) supervised machine learning methods may
be possible to use. The problem, yet, would be the fact
that there is not adequate information for sequence
similarity between existing miRNA functional
groupings which need to be experimented through
laboratory techniques. Notwithstanding, unsupervised
methods more gainful to recognize hidden relationships
between miRNAs is a fortiori in this study to use (Zhao
and Liu 2007). Hereafter, supervised clustering
techniques can be carried out and this study will be
guide for them too. Thus, this study developed a new
approach specifying the detection of miRNA sequence
groups by using various existing clustering algorithms,
we were able to instruct appropriate optimizations to
choose best possible one most fitting for miRNA
functional clustering analysis.
Statistical evaluation of clusters was completed
through DI calculations. Only the clusters significantly
showed strength of clusters used in the study. The
functional enrichments in that clusters were calculated
by very effective bioinformatics tool, Tool for
annotations of miRNA; TAM uses a given set of
miRNAs by calculating p-values of enrichment in the
set and it shows the number of sequences in the cluster
found in the same category. Our analyses have shown
the clustering approaches used in the study represent
important functional enrichments. Although, there are
some minor changes compared to TAM results when
similarity detection method changed. Most
significantly, in family category we saw the highest
enrichments indicating that sequence similarity in
miRNA families is predictable. Since, our method
yielded significant similarities it is applicable to
sequence clustering for miRNAs regardless of the small
differences that were observed in comparison to TAM
output. Thus, our results indicate that a higher
enrichment was obtained compared to any random
matrix that is used.
The final results of our analyses show that
biologically important patterns do exist in miRNA
sequences and they can be found by similarity detecting
tools. Moreover, there is important sequence
similarities in miRNAs families and this likeness can be
directly related to function due to the consequence of
the fact that miRNA family members operate together
(Burge et al. 2013). Actually, since miRNA to target
network is highly complicated (Gennarino et al. 2012),
starting from sequence similarity information may be
the first clue into functional assignment. To this end,
we suggest that the pipeline created with this study can
be used for investigations of novel miRNA datasets for
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
114

search of functional annotations. We believe that this
study will comprise a baseline for future studies.
ACKNOWLEDGEMENTS
We acknowledge Scientific and Technological Research
Council of Turkey (TÜBİTAK) for project grant under
113E527 and master thesis grant under 2210 BIDEB
program.
REFERENCES
Abbott, A.L. et al., 2005. The let-7 MicroRNA family
members mir-48, mir-84, and mir-241 function
together to regulate developmental timing in
Caenorhabditis elegans. Developmental cell, 9(3),
pp.403–14.
Altuvia, Y. et al., 2005. Clustering and conservation
patterns of human microRNAs. Nucleic acids
research, 33(8), pp.2697–706.
Antonov, A. V et al., 2009. GeneSet2miRNA: finding the
signature of cooperative miRNA activities in the gene
lists. Nucleic acids research, 37(Web Server issue),
pp.W323–8.
Asgari, S., 2011. Role of MicroRNAs in Insect Host-
Microorganism Interactions. Frontiers in physiology,
2(August), p.48.
Bartel, B. & Bartel, D.P., 2003. Update on Small RNAs
MicroRNAs : At the Root of Plant Development ? 1.
Plant physiology, 132(June), pp.709–717.
Bartel, D.P., 2013. Micro RNA Target Recognition and
Regulatory Functions. Cell, 136(2), pp.215–233.
Bartel, D.P., 2004. MicroRNAs : Genomics , Biogenesis ,
Mechanism , and Function Genomics : The miRNA
Genes. Cell, 116, pp.281–297.
Burge, S.W. et al., 2013. Rfam 11.0: 10 years of RNA
families. Nucleic acids research, 41(Database issue),
pp.D226–32.
Corpet, F., 1988. Multiple sequence alignment with
hierarchical clustering. Nucleic acids research, 16(22),
pp.10881–10890.
Dib, L. & Carbone, A., 2012. Open Access CLAG : an
unsupervised non hierarchical clustering algorithm
handling biological data.
Dopazo, J. et al., 1997. Self-organizing tree-growing
network for the classification of protein sequences.
Protein science : a publication of the Protein Society,
7(12), pp.2613–22.
Dunn, J.C., 1973. A Fuzzy Relative of the ISODATA
Process and Its Use in Detecting Compact Well-
Separated Clusters. Journal of Cybernetics, 3(3),
pp.32–57.
Dweep, H. & Gretz, N., 2015. miRWalk2.0: a
comprehensive atlas of microRNA-target interactions.
Nature methods, 12(8), p.697.
Edgar, R.C., 2010. Search and clustering orders of
magnitude faster than BLAST. Bioinformatics
(Oxford, England), 26(19), pp.2460–1.
Enright, a J., Van Dongen, S. & Ouzounis, C. a, 2002. An
efficient algorithm for large-scale detection of protein
families. Nucleic acids research, 30(7), pp.1575–84.
Flynn, P.J., 1999. Data Clustering : A Review. IEEE
Computer Society, 31(3).
Gennarino, V.A. et al., 2012. Identification of microRNA-
regulated gene networks by expression analysis of
target genes. Genome research, 22(6), pp.1163–1172.
He, L. & Hannon, G.J., 2004. MicroRNAs: small RNAs
with a big role in gene regulation. Nature reviews.
Genetics, 5(7), pp.522–31.
Herrero, J., Diaz-Uriarte, R. & Dopazo, J., 2003. Gene
expression data preprocessing. Bioinformatics, 19(5),
pp.655–656.
Herrero, J., Valencia, A. & Joaquin, D., 2001. network for
clustering gene expression patterns. , 17(2), pp.126–
136.
Hertel, J. et al., 2012. Evolution of the let-7 microRNA
Family. RNA biology, 9(3), pp.1–11.
Jacobsen, A. et al., 2013. Analysis of microRNA-target
interactions across diverse cancer types. Nature
structural & molecular biology, 20(11), pp.1325–32.
Jain, A.K., 2010. Data clustering: 50 years beyond K-
means. Pattern Recognition Letters, 31(8), pp.651–
666.
Kozomara, A. & Griffiths-Jones, S., 2011. miRBase:
integrating microRNA annotation and deep-
sequencing data. Nucleic acids research, 39(Database
issue), pp.D152–7.
Lagos-Quintana, M. et al., 2001. Identification of novel
genes coding for small expressed RNAs. Science (New
York, N.Y.), 294(5543), pp.853–8.
Lai, E.C. et al., 2003. Computational identification of
Drosophila microRNA genes. , 4(7), pp.1–20.
Li, L., Stoeckert, C.J. & Roos, D.S., 2003. OrthoMCL:
identification of ortholog groups for eukaryotic
genomes. Genome research, 13(9), pp.2178–89.
Lu, M. et al., 2008. An analysis of human microRNA and
disease associations. PloS one, 3(10), p.e3420.
Lu, M. et al., 2010. TAM: a method for enrichment and
depletion analysis of a microRNA category in a list of
microRNAs. BMC bioinformatics, 11, p.419.
Macqueen, J., 1967. Some Methods For Classification and
Analysis of Multivariate Observation. In Berkeley
Symposium on Matematical Statistic and Probablity.
University of California Press, pp. 281–297.
Needleman, S.B. & Wunsch, C.D., 1970. A general
method applicable to the search for similarities in the
amino acid sequence of two proteins. Journal of
Molecular Biology, 48(3), pp.443–453.
Newman, M.A., Thomson, J.M. & Hammond, S.M., 2008.
Lin-28 interaction with the Let-7 precursor loop
mediates regulated microRNA processing. RNA
biology, 14(8), pp.1539–1549.
Oğul, H. & Mumcuoğlu, E.U., 2007. A discriminative
method for remote homology detection based on n-
peptide compositions with reduced amino acid
alphabets. Bio Systems, 87(1), pp.75–81.
Sequence-based MicroRNA Clustering
115

Ölçer, D. & Oğul, H., 2013. Clustering MicroRNAs from
Sequence and Time-Series Expression. BIOTECHNO
2013, 5(c), pp.1–4.
Pratt, A.J. & MacRae, I.J., 2009. The RNA-induced
silencing complex: a versatile gene-silencing machine.
The Journal of biological chemistry, 284(27),
pp.17897–901.
Rawlins, T. et al., 2012. Interactive k-means clustering for
investigation of optimisation solution data. , 0, pp.1–2.
Satoh, J.-I., 2012. Molecular network analysis of human
microRNA targetome: from cancers to Alzheimer’s
disease. BioData mining, 5(1), p.17.
Shi, B., Gao, W. & Wang, J., 2012. Sequence fingerprints
of microRNA conservation. PloS one, 7(10), p.e48256.
Sisodia, D., 2012. Clustering Techniques : A Brief Survey
of Different Clustering Algorithms. International
journal of latest trends in engineering and Technlogy,
1(3), pp.82–87.
Smith, T.F. & Waterman, M.S., 1981. Identification of
common molecular subsequences. Journal of
Molecular Biology, 147(1), pp.195–197.
Zhao, D. et al., 2010. PMirP: a pre-microRNA prediction
method based on structure-sequence hybrid features.
Artificial intelligence in medicine, 49(2), pp.127–32.
Zhao, Z. & Liu, H., 2007. Spectral feature selection for
supervised and unsupervised learning. Proceedings of
the 24th international conference on Machine learning
- ICML ’07, pp.1151–1157.
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
116
