
INFERENTIAL MINING FOR RECONSTRUCTION OF 3D CELL
STRUCTURES IN ATOMIC FORCE MICROSCOPY IMAGING
Mario d’Acunto
1
, Stefano Berrettini
2
, Serena Danti
2
, Michele Lisanti
3
, Mario Petrini
4
,
Andrea Pietrabissa
5
and Ovidio Salvetti
1
1
Istituto di Scienze e Tecnologia dell’Informazione, ISTI-CNR, via Moruzzi 1, I-56124, Pisa, Italy
2
Dept. of Neuroscience, University of Pisa, via Roma 55, 56126, Pisa, Italy
3
Dept. of Orthopedics &Traumatology, University of Pisa, via Paradisa 2, 56124, Pisa, Italy
4
Dept. of Oncology & Transplants, University of Pisa, via Roma 55, 56126, Pisa, Italy
5
Dept. of Surgical Sciences, IRCCS Policlinico San Matteo, University of Pavia, 27100 Pavia, Italy
Keywords: Atomic Force Microscopy imaging, 3D cell reconstruction, Bayesian single frame high-resolution.
Abstract: Atomic Force Microscopy (AFM) is a fundamental tool for the investigation of a wide range of mechanical
properties on nanoscale due to the contact interaction between the AFM tip and the sample surface. The
focus of this paper is on an algorithm for the reconstruction of 3D stem-differentiated cell structures
extracted by typical 2D surface AFM images. The AFM images resolution is limited by the tip-sample
convolution due to the combined geometry of the probe tip and the pattern configuration of the sample. This
limited resolution limits the accuracy of the correspondent 3D image. To drop unwanted effects, we adopt
an inferential method for pre-processing single frame AFM image (low resolution image) building its super-
resolution version. Therefore the 3D reconstruction is made on animal cells using a Markov Random Field
approach for augmented voxels. The 3D reconstruction should improve unambiguous identification of cells
structures. The computation method is fast and can be applied both to multi- and to single-frame images.
1 INTRODUCTION
In this paper, we adopt an inferential procedure
providing a high-resolution algorithm for the single-
image AFM raw data and then the construction of a
3D routine for the improved resolution images.
When applied to cell populations, the 3D
reconstruction, as developed in this paper, is a useful
tool for the recognition of cell patterns and organs
and it could be used for a fast in situ analysis for
biologists and biomedical scientists.
In the last two decades, AFM has been
developed well beyond the topographic imaging
tool. It has become an important instrument for
manipulation and material property characterizations
at the nanometer scale. The precision of positioning
has always been the key driver for AFM technology
and scanning probe microscopy in general.
Nevertheless, uncontrolled hardware drift, such as
piezo creep and thermal drift, can cause image
distortion and limiting resolution. Some solutions
based on offline corrections (Yurov and Klimov,
1994), hardware optimization (Hug et al., 1992;
Altmann et al., 2000; Beyder et al., 2006), image
based real-time compensation (Clayton and Devasia,
2005), or image-based adaptive control has been
proposed (Belikov et al., 2008; D’Acunto and
Salvetti, 2011). An AFM probe tip measures the
topography of a surface by looking the vertical
deflection of a cantilever and then associating a z-
height value to the correspondent vertical deflection.
The resulting image is obtained plotting the function
z
i
=f(x
i
,y
i
), for any couple (x
i
,y
i
) of the sample
surface. The focus of this paper is to build a method
for a 3D reconstruction after the acquired AFM
image is processed in order to obtain its High-
Resolution (HR) representation.
HR methods are techniques that enhance the
resolution of an imaging system. In optical based
imaging, HR techniques break the diffraction-limit,
analogously, HR methods can improve the
resolution of digital imaging sensors. HR techniques
can be divided in two categories, single-frame or
multiple-frame, respectively. Multiple-frame HR use
the sub-pixel shifts between multiple low resolution
images of the same scene. On the contrary, single
348
D’Acunto M., Berrettini S., Danti S., Lisanti M., Petrini M., Pietrabissa A. and Salvetti O..
INFERENTIAL MINING FOR RECONSTRUCTION OF 3D CELL STRUCTURES IN ATOMIC FORCE MICROSCOPY IMAGING.
DOI: 10.5220/0003685503400345
In Proceedings of the International Conference on Knowledge Discovery and Information Retrieval (KDIR-2011), pages 340-345
ISBN: 978-989-8425-79-9
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

frame HR methods attempt to magnify the image
without introducing blur. These last methods use
other parts of the low resolution images to make an
extimation on what the high resolution images
should look like. AFM imaging requires to work
using a single-frame approach: given a single image
of sample scanned at low resolution, return the
image that is mostly likely to be generated from a
noiseless high resolution scan of the same sample
portion. After HR equilavent image is reached, 3D
recostruction is made possible using a Markov
Random Field (MRF) approach. In a MRF method
we consider a couple (h
m
, N), where a stochastic
process is indexed by an augmented voxel h
m
for
which, for every couple (x,y) of the 2D image, any
augmented voxel depends only on its immediate
neighbours of the N set, where N is a parameter
space. The choice of N depends by the system
variables conditional probability distributions, where
system variables provide the basic tool for modelling
spatial continuity.
We apply our method to cells derived by stem
primitive cells and differentiated in osteocytes or
adipocytes, or fibroblast. Any differentiated stem
cell develops specific organs and functions, and
recognition such organs is not a trivial question. The
3D reconstruction is useful when does not lose
information of the primitive image and gives the
possibility to identify unambiguous such specific
organs.
2 INFERENTIAL GENERATIVE
MODEL
Despite the AFM ability to reach high spatial
resolution, the acquired surface topography image
can sometimes not correspond to the real surface
features due to the effect of the instrument on the
object producing artefacts. These artefacts can be
generally taken into consideration while
qualitatively interpreting the AFM results. However,
3D reconstruction tools require quantitative
estimation and reconstruction of sample true
geometry. During scanning, two major AFM
artefacts can appear: a profile broadening effect due
to the tip-sample convolution and the height
lowering effect due to the elastic deformation of
studied samples.
The first effect can be schematised as follows:
the tip moving across an object surface can be
approximated by a sphere of radius R moving along
a sphere of radius r surface, i.e., the tip describes arc
of radius R+r. The lateral dimension of the surface
objects is r
c
=2(R
⋅
r)
1/2
and the relative height of the
object
Δ
z=r[1-(1-(r
c
/(R+r))
2
]
(1)
The minimum separation between two asperities or
local pattern that can be detected is d= (8RΔz)
1/2
, that
is also the lateral resolution.
Before to build the 3D structures, the source
images are processed in order to improve their
resolution. To do this operation, we adopt a
Bayesian method. Hardie et al (Hardie et al, 1997)
demonstrated that low-resolution images can be
updated using super-resolution image estimate, and
that this improves a Maximum a Posteriori (MAP)
super-resolution image estimate. Pickup et al.
(Pickup et al., 2009) used a similar joint MAP
approach to learn more general geometric patterns,
configuring the correspondent super-resolution
images and valuing the prior parameters
simultaneously. Another remarkable result for the
inferential super-resolution has been reached by
Tipping and Bishop (Tipping and Bishop, 2003),
they used a Maximum Likelihood (ML) point
estimate of the image parameters found by
integrating the high-resolution image out the
registration problem and optimising the marginal
probability of the observed low-resolution images
directly.
We follow a generative model based on an idea
as proposed by Torres-Mendez et al. (Torres-
Mendez et al., 2007) carried out from single-frame
methods. The basic idea can be summarized as
follows: given a Low Resolution (LR) image α of
size h
α
×w
α
pixels, we want to estimate the
correspondent HR image ω of size h
ω
×w
ω
, with
equal or greater size of the input image α. From α,
we must generate L images of smaller size (scaled),
that we can call observable images l, with l=0…L.
Any point in the LR image is considered as a node in
a Markovian process, and a possible neighbourhood
node in the HR image is defined by a pairwise
potential. If we denote x
i
as a set of hidden nodes in
the output ω, and the y
i
as the observable nodes in α
image, and defining the pairwise potential between
the variables x
i
, and x
j
, by
Ψ
ij
and the local evidence
potential associated with the variables x
i
and y
j
by
Φ
i
, the joint probability correspondent to the
Markovian process can be written as
()
(
)
()
ii
i
iji
ji
ij
yxxx
Z
yxP ,,
1
,
,
∏∏
= Φ
ψ
(2)
where Z is a normalization constant. Our problem
consists to maximize P(x,y), maximization that
corresponds to find the most likely state for all
hidden nodes x
i
, given all the node y
i
. To remove
INFERENTIAL MINING FOR RECONSTRUCTION OF 3D CELL STRUCTURES IN ATOMIC FORCE
MICROSCOPY IMAGING
349

ambiguities, we decide to assign high compatibility
between neighbouring pixels that have similar
intensity values and low compatibility between
neighbouring pixels that present drastic changes in
their intensity values. The value of any single pixel
in HR ω image is obtained estimating the maximum
a posteriori (MAP) solution associated to the MRF
model as given by eq. (2)
(
)
αωω
ω
P
MAP
maxarg=
(3)
where
(
)
(
)
()
()
()
ii
i
iji
ji
ij
yxxx
PPP
,,
,
∏∏
∝∝
Φ
ψ
ωωααω
(4)
Being the conditional probabilities impossible to
be exactly computed, because it is impossible to
represent all the possible combinations between
pixels, we adopt a Markov Chain Monte Carlo
(MCMC) method to approximate the best solution of
(3). The HR images so obtained are still 2D
representation of the acquired AFM images. The
correspondent 3D reconstruction is discussed in the
next section
3 3D RECONSTRUCTION
MODEL
The typical visual rendering of AFM images
describes the recorded structures assessing a gray
intensity (o colour intensity) to the z=f(x,y) measured
value. This is not properly a 3D reconstruction. 3D
reconstruction is possible in tomography-based
techniques thanks to the multi-acquisition of images
at different angles and then recollected in a unique
image via Radon anti-transformation, for example.
When the source is composed by a unique image,
the 3D reconstruction is rather complicated, and the
possibility to introduce artefacts or unwanted effects
is high. Our method is based on learning a statistical
model of the local relationship between the observed
range data and the variations in the intensity image
and uses this model to compute unknown depth
values. The intensity of any point is supposed to be a
Markov process. Unknown depth values are then
inferred by using the statistics of the observed range
data to determine the behaviour of the Markov
process. The presence of intensity where range data
is being inferred is crucial since intensity provides
knowledge of surface smoothness and variations in
depth. The advantage of our approach is to carry out
knowledge directly from the observed data, without
to introduce constraints that could be inapplicable to
particular environments. Although if our method
seem to be very close to a traditional shape-from-
shading method (where depth inference from
variations in surface shading), the substantial
difference is in the inferential engine that connects
the final 3D reconstruction to a suitable processing
of original data.
3.1 Reconstruction Methodology
Our goal is to infer a dense range map from an
intensity image and a very sparse initial range data.
The inference on range data is solved using a
sampling on the intensity at each point considered as
a product of a Markovian process. Unknown range
data is then inferred by using the statistics of the
observed range data to determine the behaviour of
the Markovian process. In our approach, there some
critical aspects, for example, the knowledge
extracted from smooth intensity variation could
generate artefacts, or again, the right weight of a
variation in depth.
The starting point is the development of a set of
augmented voxels V that contain intensity, edge
(from the intensity range) and range information. It
should be mentioned that the intensity can be
considered both for gray scales or colour images,
and that the range information includes portion of
ranges a priori unknown). Let us introduce Ω as the
area of unknown range that corresponds to the
region to be filled. Following Torres-Mendez et al.
(Torres-Mendes et al, 2007), we base our
reconstruction method on the amount of reliable
information surrounding the augmented voxel whose
depth value is to be estimated, and also on the edge
information. Thus, for each augmented voxel V
i
we
count the number of neighbour voxels with already
assigned range and intensity. A general criterion is
to start by reconstructing those augmented voxels
which have more of their neighbour voxels already
filled, leaving to the end those with an edge passing
through them. After a depth value is estimated, we
update each of its neighbours by adding 1 to their
own neighbour counters. The next step is to proceed
to the subsequent groups of augmented voxels to
synthesise until no more augmented voxels in Ω
exists.
Formally, an augmented voxel is defined as
V=(z,E,R) where z denotes the pixel intensity
directly connected to the z-height as measured by the
AFM, E is a binary matrix (1 if an edge exits, 0
otherwise) and R denotes the matrix of incomplete
pixels depth. It is possible to define a set of
KDIR 2011 - International Conference on Knowledge Discovery and Information Retrieval
350
augmented voxels that lie on each ray that intersects
each pixel of the input image z, thus giving us a
registered range image R and intensity image z. Let
h
m
=(x,y): 1≤x,y≤m denote the m integer lattice , then
z={z
x,y
}, (x,y)∈h
m
, denotes the gray levels of the
input image, and r={R
x,y
}, denotes the depth values.
Than we model the V set as a Markov Random
Field. Within the Markov Random Field picture, z
and R must be considered random variables. Let us
introduce a neighbourhood system, defined as
N={N
x,y
∈h
m
}, where N
x,y
⊂h
m
denotes the neighbours
of (x,y).
A Markov Random Field over (h
m
, N) is a
stochastic process indexed by h
m
for which, for
every couple (x,y) any augmented voxel depends
only on its immediate neighbours. The choice of N
together with the conditional probability distribution
of P(z) and P(R) provides the basic tool for
modelling spatial continuity. Therefore, the N
x,y
set
is modelled on the acquisition data matrix that is a
square mask of size n×n centered at the augmented
voxel location (x,y). The calculation of the
conditional probabilities in an explicit form is an
infeasible task since we cannot efficiently represent
or determine all the possible combinations between
augmented voxels with its associated neighbours. To
do this calculation we can invoke the Gibbs
sampling, for example, and average a depth value
from the augmented voxel V
x,y
with neighbours N
x,y
by selecting range value from the augmented
resembles the region being filled voxel whose
neighbours N
k,l
most resembles the region being
filled in
lkyx
Alk
opt
NNN
,,
),(
minarg
−=
∈
(5)
where A={A
k,l
⊂N} is the set of local neighborhood,
in which the center voxel has already assigned a
depth value, such that 1≤[(k-x)
2
+(l-y)
2
]
1/2
≤d. For
each successive augmented voxel, N
opt
as given by
Eq. (5) approximates the maximum a posteriori
estimate. The distance || ⋅ || is defined as the
weighted sum of squared differences over the partial
data in two neighbourhoods. The weights are choice
applying 2D Gaussian kernel to each
neighbourhood, such that those voxels near the
center are given more weight than those at the edge
of the window.
4 RESULTS
In this section, we present the basic results inherent
the 3D model as discussed in the past two sections.
The primitive AFM images are processed in order to
improve their quality (ranging from LR to HR) and
then the algorithm for their 3D reconstruction using
the MRF picture as given by Eq. (5) is applied.
Firstly, the method is used on images of regular
lattice for AFM calibration to sample with
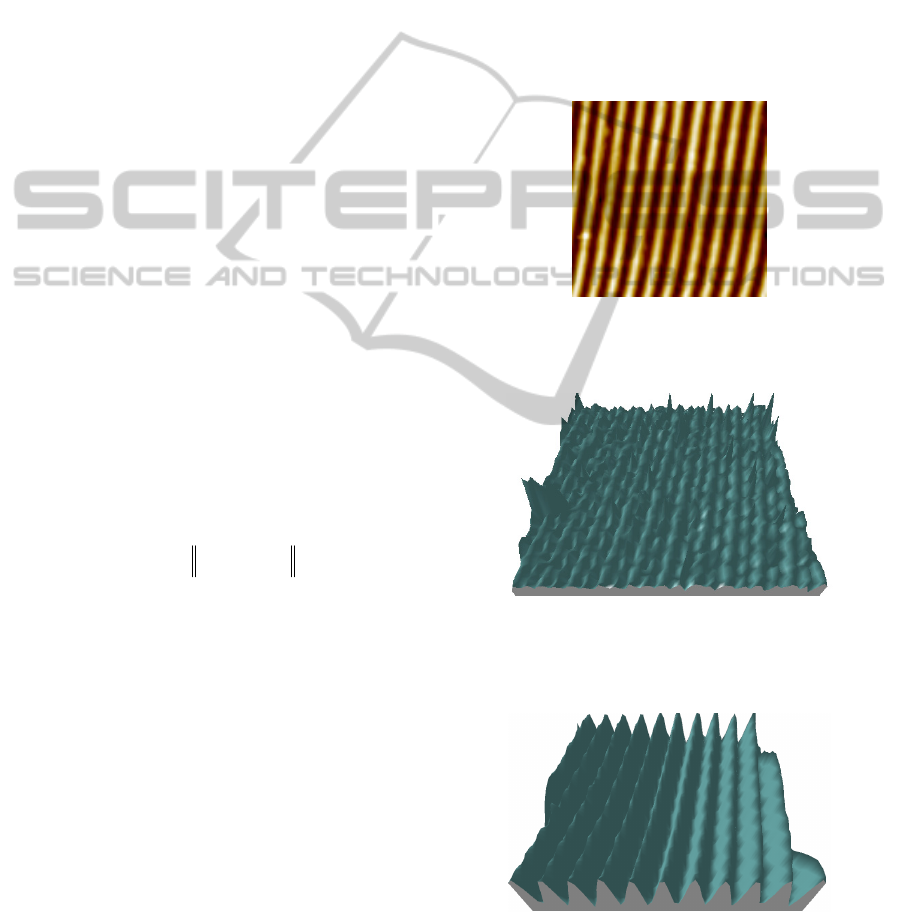
nanometers patterns. Figure 1 shows silicon grating,
normally used for calibration of z-height in AFM
measurements. Its accurate 3D reconstruction of the
grains is of great importance. It should be noted that
the image in figure 1 has not been pre-processed and
it presents an artefact at the bottom, while the image
in figure 3 that represents its correspondent 3D
reconstruction corrects such artefact.
Figure 1: Silicon grating used for AFM calibration of z-
height. The mounds are large 100nm and periodicity is
200nm.
Figure 2: 3D reconstruction of the image as in figure 1
without HR processing of the 2D image. Some artefacts
and topographic roughness present in the primitive 2D
image are amplified and the grating is not well resolved.
Figure 3: 3D reconstruction of the grating as in figure 1
with HR processing applied on the image as recorded by
the AFM. In this case, the artefacts are removed and the
grating presents a spatially well-resolved structure, where
mounds and valleys are clearly separated.
INFERENTIAL MINING FOR RECONSTRUCTION OF 3D CELL STRUCTURES IN ATOMIC FORCE
MICROSCOPY IMAGING
351
Now, we apply our procedure to cell images. The
cell samples are obtained in cellular cultures from
pluripotent stem cells and differentiated in
osteocytes, fibroblasts, adipocytes or others (Danti et
al. 2006).
A typical problem acquiring images on cells
using an AFM is the low resolution due large
dimensions of cells and reduced instrumental
capability to increase pixels. For example, many
commercial AFM can perform measurement with a
pixel density of 512×512 or 1024×1024 pixels.
Because some animal cells present dimensions that
needs scans on area of 100μm×100μm, this implies
that any single pixel covers approximately an area of
100nm×100nm for a pixel density of 1024×1024.
For this reason, the increasing of resolution can play
a fundamental role for the recognition of cells
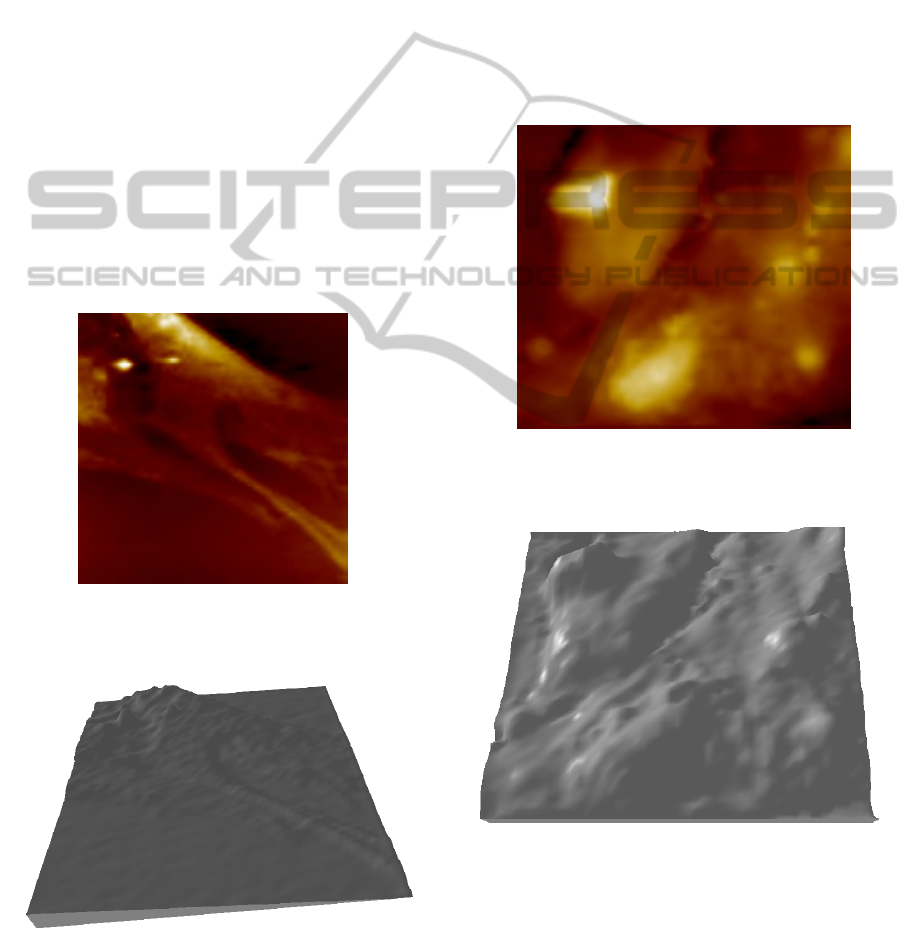
patterns or organ shapes. Figure 4 presents an image
of a large osteocyte. The primitive image is low
resolution, 512×512 pixels on an area 50μm×50μm,
after LR to HR method is applied, the 3D
reconstruction presents all the characteristics
features as in the 2D AFM image
Figure 4: AFM image of a portion of an osteocyte (real
area surface 50μm×50μm, z-height less than 5μm, density
pixel 512×512).
Figure 5: The correspondent 3D reconstruction of the
osteocyte as in figure 4.
In many cases, another problem generally found
during the imaging of cells is the identification of
specific cells in a cluster. In fact, in a cell culture,
the differentiation is often followed by meiosis, so
producing a population of cells partially overlapping
one each other. This is the case of the adipocytes
displayed in figure 6. Two nuclei are well
recognized, but it is not so for the cell edges. The 3D
reconstruction can help to identify cells dimensions
and organs in a manner that is not possible in the 2D
image and correspondent three-dimensional visual
rendering performed both with open source
(Gwyddion, http://gwyddion.net) or commercially
available programs (SPIP, http://www.imagemet.com)
Figure 6: AFM image of a cluster of adipocytes (real area
surface 50μm×50μm, z-height approximately 8μm,
density pixel 512×512).
Figure 7: 3D reconstruction of the cluster cells as in figure
6. The cells structures are well defined, it is possible to
recognize the nuclei.
5 CONCLUSIONS
Atomic Force Microscopy (AFM) is a fundamental
KDIR 2011 - International Conference on Knowledge Discovery and Information Retrieval
352

tool for the investigation of a wide range of
mechanical properties on nanoscale due to the
contact interaction between the AFM tip and the
sample surface. The information recorded with AFM
is stored and synthesized by imaging the sample
properties to be studied. The AFM topographic
images are matrices z=f(x,y), that links a z-value to
the correspondent x,y surface point. The focus of this
paper is on an algorithm for the reconstruction of 3D
structures extracted by typical 2D surface AFM
images. The AFM images resolution is limited by
the tip-sample convolution due to the combined
geometry of the probe tip, density pixels and specific
the pattern configuration of the sample. This limited
resolution reflects on the accuracy of the
correspondent 3D image. We have adopted an
inferential procedure that provides a high-resolution
algorithm for the single-image AFM raw data and
then the construction of a 3D routine for the
improved resolution images. When applied to cell
populations, the 3D reconstruction are an useful tool
for the recognition of cell pattern and organs and it
could be used for a fast in situ analysis for biologists
and biomedical scientists.
REFERENCES
Altmann S. M., Lenne P. F., Horber H., 2000. Rev. Sci.
Instrum., 72, 142-149.
Amit Y., Grenader U., Piccioni M., 1991. J. Amer. Statist.
Assoc. 86, 376-387.
Amit Y., 1994. SIAM J. Sci. Comput., 15, 207-224.
Belikov S., Shi J., Su C., 2008. American Control
conference Seattle Washington, USA, ThA08.5 (1-5).
Beyder A., Spagnoli C., Sachs F., 2006. Rev. Sci. Instrum.,
77, 1551-7.
Clayton, G. M., Devasia, S., 2005. Nanotechnology 16,
809–818.
D’Acunto M., Salvetti O., 2011, Pattern Recognition and
Image Analysis, 21, 9-19.
Danti, S., D’Acunto M., Trombi L., Berrettini S.,
Pietrabissa A., 2007. Macromolecular Bioscence, 7,
589-598.
Flores F. C., de Alecar Lotufo R., 2010. Image and Vision
Computing, 28, 1491-1514.
Gonzalez, R., Wood, R., 2008. Digital Image Processing.
Upper Saddle River, NJ, Prentice Hall.
Hardie R. C., Barnard K. J., Armstrong E. E., 1997, IEEE
Transactions on Image Processing, 6, 1621-1633.
Henn S., Witsch K., 2000. Computing, 64, 339-348.
Henn S., Witsch K., 2001. SIAM J. Sci. Comput., 23,
1077-1093.
Hug H. J., Yung T., Güntherodt H. J., 1992. Rev. Sci.
Instrum., 63, 3900-3904.
Pickup L. C., Capel D. P., Roberts S. J., Zissermann A.,
2009. The Computer Journal, 52(1), 101-113.
Tipping M. E. Bishop C. M., 2003. In Becker S. Thrun S.
and Obermeyer K. (Eds.), Advances in Neural
Information Processing Systems, 15, 1303–1310.
Torrez-Mendez L. A., Ramirez-Sosa Moran M.I., Castela
M., 2007. In Gelbuck A., Kuri Morales A. F., MICAI
2007, LNAI 2007, 640-649. Springer-Verlag Berlin
Heidelberg.
Yurov, V. Y., Klimov, A. N., 1994. Rev. Sci. Instrum.,
65(5) 1551-7.
INFERENTIAL MINING FOR RECONSTRUCTION OF 3D CELL STRUCTURES IN ATOMIC FORCE
MICROSCOPY IMAGING
353
