
RETROVIRAL GENETIC ALGORITHMS
Implementation with Tags and Validation Against Benchmark Functions
Alexander V. Spirov
1, 2
and David M. Holloway
3
1
Laboratory of Evolutionary Modelling, The Sechenov Institute of Evolutionary Physiology and Biochemistry
of the Russian Academy of Sciences, Saint-Petersburg, Russia
2
Computer Science and CEWIT, State University of New York at Stony Brook
500 Stony Brook Road, Stony Brook, NY, U.S.A.
3
Mathematics Department, British Columbia Institute of Technology, Burnaby, B.C., Canada
Keywords: Genetic algorithms, Recombination, Building blocks, Natural computing, Retroviral recombination.
Abstract: Classical understandings of biological evolution inspired creation of the entire order of Evolutionary
Computation (EC) heuristic optimization techniques. In turn, the development of EC has shown how living
organisms use biomolecular implementations of these techniques to solve particular problems in survival
and adaptation. An example of such a natural Genetic Algorithm (GA) is the way in which a higher
organism’s adaptive immune system selects antibodies and competes against its complement, the
development of antigen variability by pathogenic organisms. In our approach, we use operators that
implement the reproduction and diversification of genetic material in a manner inspired by retroviral
reproduction and a genetic-engineering technique known as DNA shuffling. We call this approach
Retroviral Genetic Algorithms, or retroGA (Spirov and Holloway, 2010). Here, we extend retroGA to
include: (1) the utilization of tags in strings; (2) the capability of the Reproduction-Crossover operator to
read these tags and interpret them as instructions; and (3), as a consequence, to use more than one
reproductive strategy. We validated the efficacy of the extended retroGA technique with benchmark tests on
concatenated trap functions and compared these with Royal Road and Royal Staircase functions.
1 INTRODUCTION
Classical understandings of biological evolution
served to inspire an entire order of heuristic
optimization techniques, known generally as
Evolutionary Computations (EC). Recent studies at
the molecular biology and genetic level have
conclusively shown that living organisms utilize
biomolecular implementations of EC for solution of
problems in survival and adaptation – for example
the selection of antibodies in a higher organism’s
adaptive immune system (Lewin, 2003) due to
competition with the development of antigen
variability in pathogenic organisms such as viruses
(Donelson, 1995); (Barbour and Restrepo, 2000).
The computational approach we present here is
inspired by the biology of retroviral reproduction, in
which genetic material is diversified through the
alternate use of DNA and RNA (Negroni and Buc,
2001; Galetto and Negroni, 2005). A virus entering a
host cell contains two or more copies of its genome
in RNA form. As part of the infection cycle, a single
DNA molecule is synthesized from the viral RNAs.
During the replication process the viral genome goes
through a series of intermediate states. Replication is
conducted by the retroviral reverse transcriptase
enzyme, which can be directed by signal elements
on the original RNA strands. Passing over these
elements during replica synthesis causes the
transcriptase to release the current template strand
and shift to a different one. These jumps (template
switches, strand transfers) are key events in
retroviral recombination (and will frequently lead to
a mutation in the replica, due to the insertion of an
extra nucleotide). The elements triggering template
switches are varied: breaks in the RNA molecule;
pause sites (RNA sequences that slow down replica
synthesis); or the local physical structure of the
RNA (e.g. a hairpin).
Computationally, the switch elements are
analogous to marks or tags on a string. Molecular
machines read these tags and interpret them as
instructions for further string operations. We use
233
V. Spirov A. and M. Holloway D..
RETROVIRAL GENETIC ALGORITHMS - Implementation with Tags and Validation Against Benchmark Functions.
DOI: 10.5220/0003674102330238
In Proceedings of the International Conference on Evolutionary Computation Theory and Applications (ECTA-2011), pages 233-238
ISBN: 978-989-8425-83-6
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

operators for reproduction following retroviral rules,
which we term Retroviral Genetic Algorithms or
retroGA (Spirov and Holloway, 2010). In the present
work, we extend retroGA to include: (1) the
utilization of tags in strings; (2) the capability of the
Reproduction-Crossover operator to read these tags
and interpret them as instructions; and (3), as a
consequence, for reproduction to use more than a
single strategy. It is commonly accepted that typical
combinatorial optimizations and biological evolution
fitness functions may be represented by rugged
landscapes. We use concatenated trap functions and
Royal Road (RR) functions to test the efficacy of the
extended retroGA approach on such landscapes.
2 THE retroGA APPROACH,
WITH TAGS
The initial implementation of retroGA (Spirov and
Holloway, 2010) included a Reproduction-Crossover
(RC) operator for the processes of retroviral
recombination. Here, we extend the RC operator by
introducing two different sets of tags that it may
operate on. This allows us to implement template
switching via signals in recombining sequences. In
addition, during rearrangement tags may be
changed, added, copied and/or moved (to another
site on the same string or to another string
altogether). This allows both the resultant string and
the processing scheme itself to change over
successive cycles of genetic rearrangement. This
enables the retroGA operators to use more than one
strategy for the recombination of parental sequences.
The RC operator: generates a child string from a
given parent pair, combining the functions of
reproduction and crossover (see Spirov and
Holloway, 2010). A pair of parents is selected, as in
standard GA, by one of several predetermined
strategies: truncation, roulette-wheel, etc. One string
is selected as a donor (tag γ), and the other is the
acceptor (tag Γ). This is analogous to retroviral
replication, with the RC operator corresponding to
reverse transcriptase and the parent strings to the
pair of retroviral RNA molecules. Like retroviruses,
replicating strings are circular (the N+1th element is
the 0th).
When the RC operator’s string reading and
copying procedure encounters a tag between the (i-
1)-th and i-th elements, it is interpreted, depending
on the nature of the tag, as one of the following
commands (tags are not copied unless explicitly
commanded, and no more than one tag is allowed
between regular string elements):
1. Finish the current child string and begin a new
one.
2. Finish the child string and terminate
reproduction.
3. Move tag one position to the right.
4. Switch template to the other parent string.
5. Replace the i-th element of the current parent or
child string with a copy of the j-th element of the
other parent string.
6. Mutate the j-th element of the donor, acceptor or
child string.
7. Insert an arbitrary element into the i-th position
of the current string.
8. Delete the j-th element from the current string.
9. Insert tag X into the j-th position of the donor,
acceptor, or child string.
More complex commands can be constructed from
these nine elementary instructions. Elementary
commands (5-9) have arguments.
Two different reproduction strategies are used: a)
replace a predetermined fraction of the population
by progeny; b) permit the operator to leave offspring
in the population if and only if their scores are
higher than their parents’.
Three-tag Model: Three tags and their respective
instructions (Table 1) are sufficient to implement
retroviral recombination in an evolutionary search
program. Processing of a pair of parent strings
begins with the insertion of replication cycle control
tags (Γ and γ). The basic operation is the generation
of a child (or replica) string, reading from left to
right, from the 0th to the Nth element, from one or
two alternating parent strings. The Γ and γ tags
model the role of viral RNA flanking regions in
controlling replication. Tag Λ is randomly inserted
into a certain fraction of the strings in the initial
population. It is composed of three elementary
instructions which together act as the entire
mechanism of retroviral recombination. The Λ tag
itself is analogous to a “stop-signal” on a retroviral
RNA molecule, in that it facilitates change of the
current template.
Table 1: The three tags and their definitions.
Tag Command
Γ Finish the current child string and begin a new
one.
γ Finish the child string and terminate reproduction.
Λ Insert random element into the i-th position of the
current parent string, delete element from the (i-
1)-th position of the current parent string, and
switch template.
ECTA 2011 - International Conference on Evolutionary Computation Theory and Applications
234
a)
1:11111111...11000000
:00000000...000000000
:11111111...11111111
child
donor
acceptor
b)
2:11111111...1000000
:00000000...000000000
:11111111...1111111
child
donor
acceptor
Figure 1: The first two recombination / replication cycles
in the 3-tag model. * indicates an arbitrary element of the
string.
Using the three tags, the RC operator creates a
complex (not ‘point’) mutation in each cycle,
inserting a random element to the right of a Λ tag,
deleting an element to the left of a Λ tag. After N
replication cycles, one Λ tag will change all N
elements of the string (see Figure 1 a, b). Without Λ
tags, the operator processes strings as in standard
GA (only point mutation and/or crossover
operators); with Λ tags, strings are processed with a
local search (C.f. the RHMC algorithm, Forrest and
Mitchell, 1993).
Eight-tag Model: Modification and broadening of
the list of tags and their corresponding commands
substantially increases the complexity of the
operator’s behaviour. We introduce an eight tag
model (Table 2) which captures the processes of
transposition (i.e. the behaviour of transposons, or
mobile genetic elements, see Spirov et al., 2009).
Tags Λ and λ are the only ones inserted randomly
into strings of the initial population. Unlike the 3-tag
model, tag Γ is now placed after the 0th element of
the acceptor string, and tag γ after the 0th element of
the donor string (Cf. Tables 1 and 2). With the
increased number of tags and commands, conflicts
may occur during tag interpretation. Specifically, a
command to move or copy a given tag X to a
position between elements (i-1) and i may result in a
collision with an already-present tag. To resolve
these conflicts, two new rules are introduced: how to
interpret an attempt to replace tag Λ with tag φ
(φ→Λ) or to replace tag φ with tag Λ (Λ→φ) (see
Table 2).
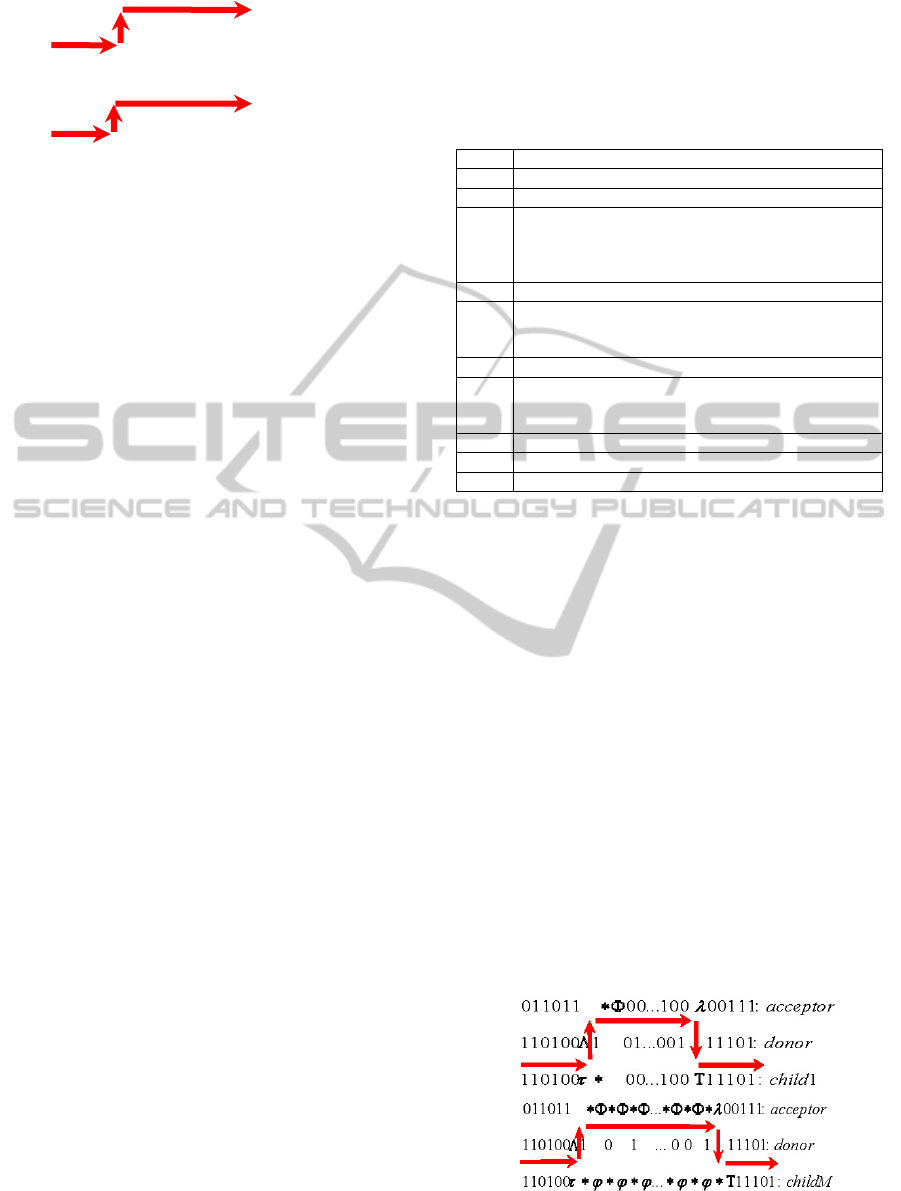
If the donor contains tag Λ in the position
between (i-1) and i, and the acceptor contains tag λ
in the position between (j-1) and j, then the region
between the i-th and j-th elements (inclusive) of the
acceptor after M recombination/replication cycles
(M = j – i) assumes the configuration
*Φ*Φ*Φ…*Φ*Φ*λ (* is an arbitrary element of the
string; see Fig. 2). Beginning with cycle M+1 the
RC operator produces progeny with random
sequences in this index range. The configuration of
this region in the child strings becomes
τ*φ*φ*φ…*φ*φ*Τ
. With this chance combination
of tags, this region of the child string can function
independently from the rest of the string.
Table 2: The eight tags and their definitions.
Tag Command
γ Finish the child string and terminate reproduction.
Γ Finish the child string and begin a new one.
λ Switch template, mutate the i-th element of the
current string, insert tag Φ into the (i+1)-th position
of the current string and insert tag τ into the child
string.
Λ Switch template and insert tag T into the child string.
φ Mutate element after the tag, copy the tag one step to
the right, insert tag φ into the same position on the
child string.
Φ Copy this tag onto the paired string.
φ→Λ Transpose tag Λ one step to the right, insert tag φ in
this position, and change the i-th element of the
current string to the i-th element of the paired string.
Λ→φ Cancel replacement, but switch template.
T Switch template.
τ Copy this tag onto the paired string.
Because of this property, a situation may arise in
a later generation where the donor string carries the
τ*φ*φ*φ…*φ*φ*Τ fragment, and the acceptor
string does not. In this case, the fragment gets copied
to the second parent (acceptor) string during a
reproductive cycle, due to combined action of tags τ,
φ, Τ (see Table 2).
If the arbitrary sequence between tags τ and Τ
forms a functional sequence (or BB, see below),
copying the fragment can be evolutionarily
favourable. By transposing itself, this fragment can
disseminate throughout the population (C.f. Spirov
et al., 2009).
GRC Operator: The fact that some retroviral
recombinatorial events can have more than two
parental sequences inspired us to generalize the RC
operator (GRC operator) to N parental strings for
each child sequence (Spirov and Holloway, 2009).
However, the GRC operator does not currently
process tags; this is one of our future directions.
a)
b)
Figure 2: The formation of a local mutagenesis mechanism
between tags Λ and λ. a) earlier; b) later.
RETROVIRAL GENETIC ALGORITHMS - Implementation with Tags and Validation Against Benchmark Functions
235

3 RESULTS AND DISCUSSION
Fitness Functions to Study Hard Evolutionary
Problems: There is every reason to believe that both
biological evolution and natural GA solve problems
of considerable difficulty. The current literature
provides grades and classifications for problem
difficulty; we select several representative types of
problem with which to benchmark our approach.
Typical combinatorial optimization or biological
evolution fitness functions may be described by
rugged landscapes (Kauffman and Levin, 1987),
with large numbers of local extrema and difficult
elements such as plateaus and valleys. Evolving
populations can typically get stuck on one of the
local peaks.
Trap Functions: Some of the simplest discrete
analogues of fitness functions with many maxima
are concatenated trap functions (Goldberg, 1987;
Goldberg, Deb, and Horn, 1992). They have been
proven to be GA hard and are of particular interest
from an experimental point of view for testing
algorithm improvements. Here we use fully
deceptive trap functions (Deb and Goldberg, 1993).
A trap function of order k is given by
F(x) = r (k-1-u(x)) / (k-1), if u(x) = k,
where u(x) counts the number of 1-bits in string x;
otherwise F(x) = 1. r<1 denotes the fitness ratio
between optimal and sub-optimal solutions. A
higher-dimensional function can be made by
concatenating n trap functions together. The bit-
string’s fitness is computed as the sum of the
fitnesses of the n traps. The concatenated trap
function has 2n local optima. The global optimum is
a string of all 1’s.
The Royal Road Fitness Functions: Mitchell and co-
workers designed a class of fitness landscapes called
Royal Road functions (RR): R1, R2, R3 and R4
(Mitchell et al., 1992; 1994); (Forrest and Mitchell,
1993). These were specifically designed to test the
“building block” (BB) approach (Goldberg, 1989);
(Holland, 1992), in which a solution can be
decomposed into BBs (which may have genetic
functional relevance), which can be searched
independently and then combined to obtain a good
or even optimal solution. RR have a fixed number of
predetermined schemata, allowing for the study of
GA performance over time. RR are a generalization
of the MaxOnes function: rather than simple
zero/one bitstrings in which the overall count of
ones determines fitness, RR strings have discrete
blocks of sub-sequences of bits, with fitness
evaluated for each block. Royal Staircase (RS) is a
variation of the Royal Road functions, using a
simple landscape with clearly defined neutral layers
(van Nimwegen and Crutchfield, 2000).
Although RR functions were designed to study
GA, some features of the RR functions, especially
R3 and R4, are reminiscent of known aspects of
molecular biological evolution (van Nimwegen and
Crutchfield, 2000, Crutchfield and van Nimwegen,
2001).
3.1 Benchmark Tests
We chose RR-type and trap functions for benchmark
performance tests of our approach versus standard
GA: they reflect many of the significant properties
of biological evolutionary searches and are well-
studied and sufficiently simple to permit statistical
analysis, allowing for comparison between
theoretical expectation and the results of
experimental runs.
The same suite of programs was used to run both
trap and RR function tests. Our package allows a
choice of either the RC or the GRC operators, and
also supports the two alternative reproduction
strategies, RStr1 and RStr2. The RC operator can
process binary strings with three tags and the
interpretation rules listed in Table 1, or eight tags
and the interpretation rules listed in Table 2. In the
current version of our package, the GRC operator is
incapable of processing tags (it ignores them). For
each of the tested fitness functions, 5 series of
experiments were performed: 3 tags and RStr1; 3
tags and RStr2; 8 tags and RStr1; 8 tags and RStr2;
and the current implementation of the GRC operator
(tags ignored). Outcomes did not depend on the
reproduction strategy.
Rugged Landscapes - Trap Fitness Functions: We
used the same parameters for trap function tests as
van Kemenade (1997). We ran a set of experiments
to characterize the efficiency of the different
approaches on different BB sizes (3, 4, 5, 6, 7 and 8
bits). The number of BBs was adjusted such that the
total length of the bit-string was approximately 40
bits. That is, starting from trap order 3, with 8192
extrema, we increased the trap function to order 8,
with 16 local extrema. We used a fitness ratio of r =
0.7.
In all runs, the search terminated when the
optimal solution was obtained, or when the number
of function evaluations exceeded 500,000. The
initial population size was 4096 strings. All results
(Fig. 3) are averaged over 1000 independent runs.
As seen in Fig. 3, the 3-tag version of the retroGA
ECTA 2011 - International Conference on Evolutionary Computation Theory and Applications
236
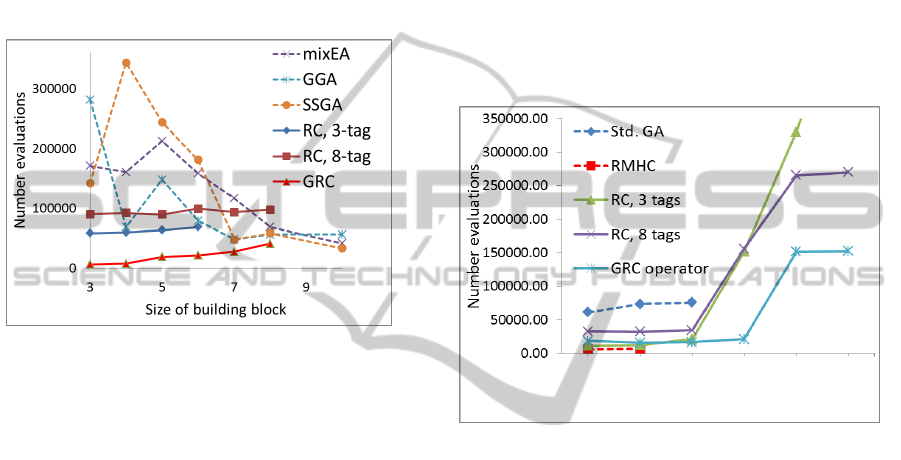
operator is more efficient than the algorithms
(including ‘general’ or standard GA, GGA)
developed by van Kemenade (1997), while the 8-tag
results are not so impressive. To our surprise, the
best performance was achieved by the GRC operator
– some 20 to 50 times faster than standard GA, on
average, for order-3 and order-4 trap functions. We
conclude that elaborate consensus-dependent
operators (standard GA) are not as effective on
rugged landscapes as the more straightforward GRC
operator.
Figure 3: Performance of our approach versus standard
GA. Values indicate number of function evaluations
needed to reach optimum. The results on the mixEA, GGA
and SSGA algorithms are from van Kemenade (1997).
Subbasin-portal Architecture - The RR Fitness
Functions: We tested our approaches on four RR
functions (R1-R4), highlighting different levels of
efficiency to the different functions (Fig. 4). RC, 3-
tag was very efficient for R1-R3, but did not reach
R4-5th level or RS. RC, 8-tag was very impressive
for solving all test functions. In R1-R4, this
approach outperformed standard GA: it was twice as
effective in R1, and even more so in R2 and R3. R4
is well-known to be hard to reach for many
optimization approaches, both evolutionary and non-
evolutionary (Mitchell et al., 1992); (Forrest and
Mitchell, 1993); (Mitchell et al., 1994). The RC, 8-
tag approach, however, achieved the fourth level of
R4 in approximately 30% of runs. Neither Standard
GA, nor Random-Mutation Hill-Climbing (RMHC)
reached the 4th (or 5th) level within the maximum of
10
6
function evaluations (Forrest and Mitchell,
1993). In 4% of cases, RC, 8-tag reached the fifth
level of the R4 test, a success rate unprecedented in
the EC literature.
Surprisingly, it was the GRC operator that ended
up being the most effective of all the strategies
tested. Notably, its performance on R1 approached
the non-evolutionary RMHC algorithm (which is not
successful on the higher test functions). For R1, it
was only three times less effective than RMHC (or
even two times, depending on operator parameters),
while RMHC outperformed standard GA by a factor
of 10. The GRC operator achieved the fourth level
of the R4 test in 98% of the runs, and the fifth level
in 67%. With the GRC operator, we have found an
evolutionary approach that outperforms standard GA
by a factor of 3 to 4 on all RR functions. GRC
operator success rates on R4 were unprecedented.
For the RS test function, the 8-tag strategy found the
answer twice as fast, on average, than standard GA,
while the GRC operator was more than three times
faster than GA (Fig. 4).
Figure 4: Performance of our approaches (RC & GRC
operators) versus standard GA (Std. GA) and Random-
Mutation Hill-Climbing (RMHC) on the Royal Road
family functions. Values indicate number of function
evaluations needed to reach optimum, averaged over 1000
runs. R4
L4
and R4
L5
are the 4
th
and the 5
th
level of the R4,
respectively. It takes >500,000 evaluation to solve the RS
problem by Std. GA and RC, 3-tag.
Comparison of Figs. 3 and 4 indicates that the
retroGA-with-tags approaches are more effective on
subbasin-portal functions (RR-type, versus trap
functions). We can hypothesize that these models
have picked up some of the crucial features of real
molecular recombinatorial mechanisms which
operate within such architectures.
We conclude that there is a fundamental
difference in the quality of artificial recombination
implemented by the GRC operator and by the
standard GA crossover operator. The positions of the
sites of crossover and exchange between two strings
in computational GA are chosen randomly.
However, in biology, crossover occurs at sites of
high homology between two molecules of nucleic
acid. These regions of high homology may be
naturally interpreted as BBs. As such, crossover
R1 R2 R3 R4
L4
R4
L5
RS
RETROVIRAL GENETIC ALGORITHMS - Implementation with Tags and Validation Against Benchmark Functions
237

operations in the natural world do not destroy BBs,
but instead conserve them wholly; it is the material
between the BBs that undergoes crossover
exchanges and point mutations. It is well-known that
the destruction of already-discovered BBs by
crossover operators is one of the major problems
with standard GA (originally shown through
experiments with RR functions). Because of this, the
ability of homology-based mechanisms (e.g. sex-
based polymerase chain reaction) to conserve
already located BBs is of tremendous interest to us.
The longer-term goals of our project are to
develop the retroGA approaches such that we can
more clearly gauge their utility to computer science
in general, as well as in such practical applications
as in vitro molecular evolution and biomolecular
computation. In recent decades, computational GA
has become an effective mathematical instrument for
modelling and analyzing the processes and
mechanisms of biological evolution. As retroGA is
for the most part domain-independent, it can readily
be applied to all forms of EC, for example greatly
assisting in solving problems on the selection of
macromolecules with properties that do not exist in
the natural world.
ACKNOWLEDGEMENTS
This work was supported by Joint NSF/NIGMS
BioMath Program, 1-R01-GM072022 and the
National Institutes of Health, 2R56GM072022-06.
REFERENCES
Barbour A. G., and Restrepo B. I., (2000). Antigenic
variation in vector-borne pathogens. Emerg Infect Dis.
6: 449-457.
Crutchfield, J. P. and van Nimwegen, E., (2001). The
Evolutionary Unfolding of Complexity. In Evolution
as Computation, DIMACS workshop, Springer-Verlag,
New York.
Deb, K. and Goldberg, D. E., (1993). Analyzing deception
in trap functions In D. Whitley (Ed.), Foundations of
Genetic Algorithms, pp. 93-108.
Donelson J.E. (1995). Related Mechanisms of antigenic
variation in Borrelia hermsii and African
trypanosomes. J Biol Chem. 270:7783-7786.
Forrest S. and Mitchell M., (1993) Relative building-block
fitness and the buildingblock hypothesis. In D.
Whitley (ed.), Foundations of Genetic Algorithms 2,
109-126. San Mateo, CA: Morgan Kaufmann.
Galetto R. and Negroni M., (2005), Mechanistic features
of recombination in HIV. AIDS reviews 7 (2: 92-102.
Goldberg D. E., (1987). Simple genetic algorithms and the
minimal deceptive problem. In L. Davis, editor,
Genetic Algorithms and Simulated Annealing, pp. 74-
88. Pitman, London.
Goldberg, D. E., (1989). Genetic Algorithms in Search,
Optimization, and Machine Learning. Addison-
Wesley, Reading, Massachusetts.
Goldberg D. E., Deb K., and Horn J., (1992). Massive
multimodality, deception, and genetic algorithms, In:
Parallel Problem Solving from Nature, 2, pp. 37-46.
Holland, J. H., (1992). Adaptation in natural and Artificial
Systems: an introductory analysis with applications to
biology, control and artificial intelligence. MIT Press.
Kauffman S. A. and Levin S., (1987). Towards a general
theory of adaptive walks on rugged landscapes. J.
Theor. Biol., 123:11-45.
Kemenade C. H. M., van, (1997). The Mixing
Evolutionary Algorithm, independent selection and
allocation of trials. In Proceedings of the IEEE
international conference on evolutionary computation,
1997, p. 13-18.
Lewin, B. (2003). Genes VIII. 1056 p.
Mitchell M., Forrest S., and Holland J. H., (1992). The
Royal Road for genetic algorithms: Fitness landscapes
and GA performance. In Proceedings of the First
European Conference on Artificial Life. Cambridge,
MA: MIT Press/Bradford Books.
Mitchell M., Holland J., and Forrest S., (1994) When Will
a Genetic Algorithm Outperform Hill Climbing? In J.
Cowan, G. Tesauro, and J. Alspector, Advances in
Neural Information Processing Systems, Morgan
Kauffman, San Francisco, CA.
Negroni M. and Buc H., (2001). Mechanisms of retroviral
recombination. Annu Rev Genet. 35: 275-302.
Nimwegen E, van, and Crutchfield, J. P., (2000).
Metastable Evolutionary Dynamics: Crossing Fitness
Barriers or Escaping via Neutral Paths? Bulletin of
Mathematical Biology 62: 799-848.
Spirov A. V., Kazansky A. B., Zamdborg L., Merelo J. J.,
Levchenko V. F., (2009), Forced Evolution in Silico
by Artificial Transposons and their Genetic Operators:
The John Muir Ant Problem CoRR abs/0910.5542
Spirov A. V. and Holloway D. M., (2010) Design of a
dynamic model of genes with multiple autonomous
regulatory modules by evolutionary computations.
Procedia Computer Science 1(1): 1005-1014.
ECTA 2011 - International Conference on Evolutionary Computation Theory and Applications
238
