
EVALUATING PERFORMANCE OPTIMIZATIONS OF
LARGE-SCALE GENOMIC SEQUENCE SEARCH APPLICATIONS
USING SST/MACRO
Tae-Hyuk Ahn
1
, Damian Dechev
2,3
, Heshan Lin
1
, Helgi Adalsteinsson
3
and Curtis Janssen
3
1
Department of Computer Science, Virginia Tech, VA 24061, Blacksburg, U.S.A.
2
Department of Electrical Engineering and Computer Science, University of Central Florida, FL 32816, Orlando, U.S.A.
3
Scalable Computing R&D Department, Sandia National Laboratories, CA 94551, Livermore, U.S.A.
Keywords:
Exascale architecture simulator, mpiBLAST, Performance and scalability modeling.
Abstract:
The next decade will see a rapid evolution of HPC node architectures as power and cooling constraints are
limiting increases in microprocessor clock speeds and constraining data movement. Future and current HPC
applications will have to change and adapt as node architectures evolve. The application of advanced cycle
accurate node architecture simulators will play a crucial role for the design and optimization of future data
intensive applications. In this paper, we present our simulation-based framework for analyzing the scalability
and performance of a number of critical optimizations of a massively parallel genomic search application,
mpiBLAST, using an advanced macroscale simulator (SST/macro). In this paper we report the use of our
framework for the evaluation of three potential improvements of mpiBLAST: enabling high-performance par-
allel output, an approach for caching database fragments in memory, and a methodology for pre-distributing
database segments. In our experimental setup, we performed query sequence matching on the genome of the
yellow fever mosquito, Aedes aegypti.
1 INTRODUCTION
The exponential growth of data intensive applica-
tions and the necessity for complex and massive
data analysis have elevated modern large-scale par-
allel computing technology and demand. Future
High-Performance Computing (HPC) systems will go
through a rapid evolution of node architectures as
power and cooling constraints are limiting increases
in microprocessor clock speeds. Consequently com-
puter architects are trying to increase significantly the
on-chip parallelism to keep up with the demands for
fast performance and high volume of data process-
ing. Multiple cores on a chip is no longer cutting edge
technologydue to this hardware paradigm shift. In the
new Top 500 supercomputer list published in March
2011, more than 99% of supercomputers are multi-
core processors (Top500, 2011). As hardware has
evolved, software applications must adapt and gain
the capability of effectively running multiple tasks si-
multaneously through parallel methods. It is of criti-
cal importance to provide an accurate estimate of an
application’s performance in a massively parallel sys-
tem both for predicting the most effective design of a
multi-core large-scale architecture as well as for op-
timizing and fine-tuning the software application to
efficiently execute in such a highly concurrent envi-
ronment.
A key element of the strategy as we move for-
ward is the co-design of applications, architectures,
and programming environments, to navigate the in-
creasingly daunting constraint-space for feasible ex-
ascale system design. The complexity of designing
large-scale computer systems has motivated the de-
velopment and utilization of a large number of cycle-
accurate hardware and system simulators (Janssen
et al., 2010; Underwood et al., 2007; Sherwood et al.,
2002). There is a pressing need to develop accu-
rate code analysis and system simulation platforms
to insert application developers directly into the de-
sign process for HPC systems in the exascale era. It
is of significant importance to build the simulation
platforms for accurate emulation of the hardware ar-
chitectures of the next decade and their design con-
65
Ahn T., Dechev D., Lin H., Adalsteinsson H. and Janssen C..
EVALUATING PERFORMANCE OPTIMIZATIONS OF LARGE-SCALE GENOMIC SEQUENCE SEARCH APPLICATIONS USING SST/MACRO.
DOI: 10.5220/0003600200650073
In Proceedings of 1st International Conference on Simulation and Modeling Methodologies, Technologies and Applications (SIMULTECH-2011), pages
65-73
ISBN: 978-989-8425-78-2
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

Application Threads Discrete Event Simulator
(Lightweight Threads)
P roces s
C a llbacks
E vents
E vents
C a llbacks
S imulatorInterface
K ernels
S ervers
R eques t
T race
S keleton
C P U
Network
Figure 1: SST/macro Simulation Framework.
straints. This will enable computer scientists to en-
gage early in the design and utilization of effective
programming models.
Three well-known approaches have been inves-
tigated for estimating large-scale performance. The
most common approach is direct execution of the full
application on the target system (Prakash et al., 2000;
Riesen, 2006; Zheng et al., 2005). This simulation
approach uses virtual time unlike normal benchmark-
ing that uses real time. Here, performance is modeled
by using a processor model and communication work
in addition to simulated time for a modeled network.
Another approach is tracing the program in order to
collect information about how it communicates and
executes (Zheng et al., 2005). The resulting trace file
contains computation time and actual network traffic.
Tracing provides high levels of evaluation accuracy,
but cannot be easily scaled to a different number of
processors. A third approach is to implement a model
skeleton program that is a simple curtailed version of
the full application but provides enough information
to simulate realistic activity (Susukita et al., 2008;
Adve et al., 2002). This approach has the advantage
that the bulk of the computational complexity can be
replaced by simple calls with statistical timing infor-
mation. What makes this approach challenging is the
necessity to develop a model skeleton program based
on a complex scientific HPC application that often in-
cludes a large number of HPC computational methods
and libraries, sophisticated communication and syn-
chronization patterns, and architecture-specific opti-
mizations. Moreover, it is difficult to analyze and
predict the runtime statistics for domain-specific ap-
plications using heuristic algorithms. The skeleton
application provides a powerful method for evaluat-
ing the scalability and efficiency over various archi-
tectures of moderate or extreme scales. For example
by running skeleton applications, the Structural Sim-
ulation Toolkit’s macroscale simulator (SST/macro)
(Janssen et al., 2010; SST/Macro, 2011) has been able
to model application performance at levels of paral-
lelism that are not obtainable on any known existing
HPC system.
In this work we present the design and application
of a discrete event simulation-based framework for
analyzing the scalability and performance of a num-
ber of optimizations of mpiBLAST. mpiBLAST (Dar-
ling et al., 2003) is an open-source parallel imple-
mentation of the National Center for Biotechnology
Information’s (NCBI) Basic Local Alignment Search
Tool (BLAST) (Altschul et al., 1990). BLAST is the
most widely used genomic sequence alignment algo-
rithm. Though a heuristic method is employed to im-
prove computational efficiency, computation time is
debilitating because of the rapid growth of sequence
data. A parallel version of BLAST, mpiBLAST, uses
a database segment approach. The design of mpi-
BLAST has been revised a number of times to better
address the challenges of distributed result processing
(Lin et al., 2005), hierarchical architectures (Thorsen
et al., 2007), include further dynamic load balancing
optimizations, and I/O optimizations (Lin et al., 2008;
Lin et al., 2011). Our simulation-based framework
allows programmers to better address the challenges
of executing genomic sequence alignment algorithms
on many-core architectures and at the same time gain
important insights regarding the effectiveness of the
mentioned mpiBLAST optimization techniques. This
allows both scientists, library developers, and hard-
ware architecture designers to evaluate the scalability
and performance of a data intensive application on a
wide variety of multi-core architectures, ranging from
a regular cluster machine to a future many-core petas-
cale supercomputer. Our approach can help in several
ways including:
• enhance the evolution of the software application
by performing further architecture-specific opti-
mizations to meet the challenges of the communi-
cation and synchronization bottlenecks of the new
multiprocessor architectures,
SIMULTECH 2011 - 1st International Conference on Simulation and Modeling Methodologies, Technologies and
Applications
66

• adapt the hardware set-up to better facilitate the
computational and communication patterns of the
application,
• evaluate the effectiveness and associated trade-
offs of any future co-design evolution of the ap-
plication software and the hardware platform.
In this paper, we present the application of SST/-
macro, an event-driven cycle-accurate macroscale
simulator, for estimating and predicting the perfor-
mance of large-scale parallel bioinformatics applica-
tions based on mpiBLAST. SST/macro has been re-
cently developed and released by the Scalable Com-
puting R&D Department at Sandia National Labs and
is a fully component-based open source project that
is freely available to the research and academic com-
munity (SST/Macro, 2011). We demonstrate the use
of SST/macro and its trace-driven simulation that is
based on DUMPI (Janssen et al., 2010), a custom-
built MPI tracing library developed as a part of the
SST/macro simulator. We also present a methodology
for constructing SST/macro skeleton programs based
on mpiBLAST.
The rest of this work is organized as follows: Sec-
tion 2 introduces the event-driven SST/macro sim-
ulator that is at the core of our simulation frame-
work, Section 3 discusses the mpiBLAST algorithm
for parallel genome sequence matching and the pos-
sible optimizations of mpiBLAST, Section 4 presents
in details the methodology of collecting DUMPI trace
files and our approach for implementing mpiBLAST-
based skeleton models as well as our experimental
set-up and results, and Section 5 concludes this pa-
per.
2 EVENT-DRIVEN
MACROSCALE SIMULATION
We begin with a discussion of the high-level design
and functionality of the SST macroscale simulator
that is at the core of our framework. The overall net-
work topology and model are presented in brief. Then
we discuss MPI modeling through skeleton applica-
tions and MPI trace files.
The purpose of a large number of simulation
tools and strategies is to help design new hardware
platforms and better applications in HPC comput-
ing. The macroscale version of the Structural Simula-
tion Toolkit is an architectural simulator that permits
coarse-grained study of data intensive parallel scien-
tific applications. SST/macro has a modular struc-
ture implemented in C++ (Stroustrup, 2000), allow-
ing flexible addition of new components and modifi-
cations. Figure 1 shows the highlight of the design of
the SST/macro simulator. The simulator makes use
of extremely lightweight application threads, allow-
ing it to maintain simultaneous task counts ranging
into the millions. Task threads create communication
and compute kernels, then interact with the simula-
tor’s back-end by pushing kernels down to the inter-
face layer. The interface layer generates simulation
events and handles the scheduling of resulting events
to the simulator back-end. The interface layer imple-
ments servers to manage the interaction with the net-
work model in the context of the application. SST/-
macro supports two execution modes: skeleton ap-
plication execution and trace-drivensimulation mode.
The processor layer receives callbacks when the ker-
nels are completed.
Recent growth of large-scale systems has made
evaluation of communication loads across complex
networks vital. SST/macro is capable of simulating
and evaluating advanced network workload with di-
verse topology and routing. The simulator currently
supports torus, fat-free, hypercube, Clos, and gamma
topologies, all described further in (Dally and Towles,
2003). Moreover, the general frameworkof a network
can be easily evaluated with network parameters such
as bandwidth and latency, thus allowing the capture
of actual trade-offs between fidelity and runtime of a
system’s network. The routing algorithms are static in
SST/macro, i.e., messages between two processors al-
ways follow the same path regardless of network sta-
tus. The modularity of the simulator makes defining
new connections easy.
2.1 The MPI Model
The Message Passing Interface (MPI) is a message
passing library interface specification for a distributed
parallel memory system (MPI, 2009). MPI primarily
allows message-passing communication from the ad-
dress space of one process to that of another process.
MPI is not a language, and all MPI operations are
expressed as functions, subroutines, or methods, ac-
cording to the appropriate language bindings, for C,
C++, and FORTRAN. The main advantages of MPI
are portability and usability. The standard includes
two main privileges: point-to-point message passing
and collective operations. A number of important
MPI functions involve communication between two
specific processes based on point-to-point operations.
MPI specifies mechanisms for both blocking and non-
blocking point-to-point communication mechanisms.
A procedure is blocking if returning from the proce-
dure indicates the user is allowed to reuse resources
specified in the call and a procedure is nonblocking if
EVALUATING PERFORMANCE OPTIMIZATIONS OF LARGE-SCALE GENOMIC SEQUENCE SEARCH
APPLICATIONS USING SST/MACRO
67

the procedure may return before the operation com-
pletes. Collective operation is defined as communica-
tion that involves groups of processors to invoke the
procedure. These include such operations as all-to-all
(all processes contribute and receive the result), all-
to-one (all processes contribute to the result and one
process receives the result), and one-to-all (one pro-
cess contributes to the result and all processes receive
the result).
In the SST/macro simulator, lightweight applica-
tion threads perform MPI operations. SST/macro im-
plements a complete simulated MPI which skeleton
applications can use to emulate node communication
in a direct manner. SST/macro has been used to test
the performance impact of proposed extensions to the
MPI standard. A simple processor model is added
to provide timings for processor workload and data
movement within each node. SST includes a network
layer that supports a large array of interconnects. The
application trace and CPU model helps to determine
when a computation operation completes and sched-
ules a completion event. The SST/macro components
provide a complete performance estimation environ-
ment for HPC platforms.
2.2 Trace File Simulation
In the trace file simulation approach, an application
is executed and profiled in order to extract a wealth
of information about its execution pattern such as the
average instruction mix, memory access patterns, and
communication mechanisms and bottlenecks. The
network utilization on a per-link basis is also esti-
mated. The generated trace file contains data such
as the time spent in computation and the commu-
nication footprints between processors. SST/macro
supports two trace file formats: Open Trace Format
(OTF) (Kn¨upfer et al., 2006) and DUMPI (Janssen
et al., 2010). OTF is a trace definition and representa-
tion format designed for use with large-scale parallel
platforms. The authors in (Kn¨upfer et al., 2006) iden-
tify three main design goals of OTF: openness, flex-
ibility, and performance. DUMPI is a custom trace
format developed as a part of the SST/macro simula-
tor. Both of the trace formats record execution infor-
mation by linking the target application with a library
that uses the PMPI (Mintchev and Getov, 1997) in-
terface to intercept MPI calls. The DUMPI format
is designed to record more detailed information com-
pared to OTF, including the full signature of all MPI-1
and MPI-2 calls. In addtion, DUMPI trace files store
information regarding the return values and the MPI
requests, which allows error checking and MPI op-
eration matching. DUMPI files also provide proces-
sor hardware performance counter information using
the Performance Application Programming Interface
(PAPI) (Janssen et al., 2010), which allows informa-
tion such as cache misses and floating point opera-
tions to be logged.
2.3 Skeleton Application Simulation
The main advantage of trace file driven simulation is
accuracy, especially if the planned runtime system is
known in details. However, a main difficulty is the
fact that it requires the execution of the actual appli-
cation that could often be data intensive and of high
computational complexity. Moreover, trace file sim-
ulation is not capable of predicting performance on
future hardware platforms, as the generated trace files
are specific to the execution environment.
Skeleton applications are simplified models of ac-
tual HPC programs with enough communication and
computation information to simulate the application’s
behavior. One method of implementing a skeleton ap-
plication is to replace portions of the code performing
computations with system calls that instruct the sim-
ulator to account for the time implicitly. Since the
performance models can be embedded in the skeleton
application and real calculations are not performed,
the simulator requires significantly less computational
cost than simulating the entire system. Skeleton ap-
plication simulation can also evaluate efficiency and
scalability at extremely different scales, which pro-
vides a powerful option for performance prediction
of non-existing super-scalar systems. Though driving
the simulator with a skeleton application is a pow-
erful approach for evaluating the application’s scal-
ability and efficiency, it requires extensive efforts for
programmers to implement the skeleton models for
a large-scale parallel program. The effort is justi-
fied by the difficulty of predicting computation time
for complex applications such as mpiBLAST. mpi-
BLAST search time varies greatly across the same
size database and query, because the computation
time depends on the number of positive matches
found in a query. Match location can also affect the
execution time.
Figure 2 shows the implementation of an MPI
ping-pong skeleton application in which pairwise
ranks communicate with each other. As shown in Fig-
ure 2, skeleton application implementations for the
SST/macro are very similar to the native MPI im-
plementation with the exception of the syntax of the
MPI calls. In addition to replacing the communica-
tion calls, we can replace computation parts with sys-
tem calls such as compute(...), which reduce simula-
tion time dramatically.
SIMULTECH 2011 - 1st International Conference on Simulation and Modeling Methodologies, Technologies and
Applications
68

void
mpipingpong ::run() {
timestamp start = mpi()->init();
mpicomm world = mpi()->comm_world();
mpitype type = mpitype::mpi_double;
int
rank = world.rank().id;
int
size = world.size().id;
/ / With an odd number of nodes ,
/ / rank ( size −1) s i t s out
if
(!((size % 2)&&(rank+1 >= size))){
/ / partner nodes 0<=>1, 2<=>3, etc .
mpiid peer(rank ˆ 1);
mpiapi::const_mpistatus_t stat;
for
(
int
half_cycle = 0;
half_cycle < 2 * num_iter;
++half_cycle) {
if
((half_cycle + rank) & 1)
mpi()->send(count, type , peer ,
tag, world);
else
mpi()->recv(count, type , peer ,
tag, world , stat);
}
}
timestamp done = mpi()->finalize();
}
Figure 2: Core execution loop of the MPI ping-pong skele-
ton application.
3 mpiBLAST
This section lays out the core design and function-
ality of mpiBLAST. Furthermore, we discuss the I/O
and computation scheduling optimization proposed to
mpiBLAST.
3.1 The Fundamental Design of
mpiBLAST
In bioinformatics, a sequence alignment is an es-
sential mechanism for the discovery of evolutionary
relationships between sequences. One of the most
widely used alignment search algorithms is BLAST
(Basic Local Alignment Search Tool) (Altschul et al.,
1990; Altschul et al., 1997). The BLAST algorithm
searches for similarities between a set of query se-
quences and large databases of protein or nucleotide
sequences. The BLAST algorithm is a heuristic
search method for finding locally optimal alignments
or HSP (high scoring pair) with a score of at least
the specified threshold. The algorithm seeks words of
length W that score at least T when aligned with the
query and scored with a substitution matrix. Words
in the database that score T or greater are extended
in both directions in an attempt to find a locally opti-
mal un-gapped alignment or HSP (high scoring pair)
with a score of at least E value lower than the spec-
ified threshold. HSPs that meet these criteria will be
reported, provided they do not exceed the cutoff value
specified for the number of descriptions and/or align-
ments to report.
Today, the number of stored genomic sequences
is increasing dramatically, which demands higher
parallelization of sequence alignment tools. More-
over, next-generation sequencing, a new generation of
non-Sanger-based sequencing technologies, has pre-
sented new challenges and opportunities in data inten-
sive computing (Schuster, 2007). Many parallel ap-
proaches for BLAST have been investigated (Braun
et al., 2001; Bjornson et al., 2002; Mathog, 2003;
Lin et al., 2005), and mpiBLAST is an open-source,
widely used parallel implementation of the NCBI
BLAST toolkit.
The original design of mpiBLAST follows a
database segmentation approach with a master/-
worker system. It works by initially dividing up
the database into multiple fragments. This pre-
processing step is called
mpiformatdb
. The mas-
ter uses a greedy algorithm to assign and distribute
pre-partitioned database chunks to worker processors.
Each worker then concurrently performs a BLAST
search on its assigned database fragment in paral-
lel. The master server receives the results from each
worker, merges them, and writes the output file. mpi-
BLAST achieves an effective speedup when the num-
ber of processors is small or moderate. However, mpi-
BLAST suffers from non-search overheads when the
number of processors increases and the database size
varies. Additionally, the centralized output process-
ing design can greatly hamper the scalability of mpi-
BLAST.
3.2 Optimizations of mpiBLAST
Hierarchical Architecture. mpiBLAST expands
the original master-worker design to hierarchical de-
sign, which organizes all processes into equal-sized
partitions by a supermaster process. The supermas-
ter process manages assigning tasks to different par-
titions and handling inter-partition load balancing.
There is one master processor for each partition that
is responsible for coordinating both computation and
I/O scheduling with many workers in a partition. This
hierarchical design has an advantage in massive-scale
parallel machines as it distributes the workload well
across multiple partitions.
Dynamic Load Balancing Design. It is difficult to
estimate the execution time of BLAST because search
time is extremely variable and thus unpredictable
EVALUATING PERFORMANCE OPTIMIZATIONS OF LARGE-SCALE GENOMIC SEQUENCE SEARCH
APPLICATIONS USING SST/MACRO
69

(Gardner et al., 2006). Therefore a greedy scheduling
algorithm for fine-grained task assignment to idle pro-
cesses is necessary. To avoid load imbalance while re-
ducing the scheduling overhead, mpiBLAST adopts a
dynamic worker group management approach where
the masters dynamically maintain a window of out-
standing tasks. Whenever a worker finishes its tasks,
it requests further assignments from its master. With
query prefetching, the master requests the next query
segment when the total number of outstanding tasks
in the window falls under a certain threshold.
Parallel I/O Strategy. Massive data I/O can lead
to performance bottlenecks especially for data driven
applications such as mpiBLAST. To deal with this
challenge, mpiBLAST pre-distributes database frag-
ments to workers before the search begins. Workers
cache database fragments in memory instead of lo-
cal storage. This is recommended on diskless plat-
forms where there is no local storage attached to each
processor. By default, mpiBLAST uses the master
process to collect and write results within a parti-
tion, which may not be suitable for massively paral-
lel sequential search. Asynchronous parallel output
writing techniques optimize concurrent noncontigu-
ous output access without inducing synchronization
overhead which result from traditional collective out-
put techniques.
4 EXPERIMENTAL RESULTS
We chose to identify and use freely available datasets
for executing our mpiBLAST-based simulation anal-
ysis. In our experimental set-up we run mpiBLAST
on the genome of the yellow fever mosquito, Aedes
aegypti, which has been investigated by biologists
for spreading dengue and yellow fever viruses. The
genome database can be downloaded freely from the
source in (Vectorbase, 2010) and has a suitable size
of 1.4GB for testing on both our local machine and
the cluster system. We use 1MB sequences randomly
sampled from the Aedes aegypti transcriptome dataset
because such query sequences match well with the
genome’s characteristics.
In our experiments, we relied on DUMPI to fa-
cilitate more detailed tracing of MPI calls than was
available from other trace programs. The results of
a DUMPI profiling run consists of two file formats.
One is an ASCII metafile for the entire run, and the
other is a binary trace file for each node. The metafile
is a simple key/value ASCII file that is intended to
be human-readable and to facilitate grouping related
trace files together. Each trace file consists of a 64-
bit lead-in magic number and 8 data records. In order
to trace an application with DUMPI, a collection of
DUMPI libraries are linked to the application when
it is executed in the system. Afterwards, several exe-
cutables built on DUMPI repository are used to ana-
lyze the DUMPI trace files.
In our experimental setup we have traced and an-
alyzed the mpiBLAST implementation described in
Section 3. The current open-source version of mpi-
BLAST has several options for parallel input/ouput
of data. We have simulated and tested three optimiza-
tions as described below:
• Optimization 1 (--use-parallel-write), enabling high-
performance parallel output: by default, mpiBLAST
uses the master process to collect and write results
within a partition. This is the most portable output
solution and should work on any file system. How-
ever, using the parallel-write solution is highly rec-
ommended on platforms with fast network intercon-
nection and high-throughput shared file systems.
• Optimization 2 (--use-virtual-frags), enabling work-
ers to cache database fragments in memory instead
of on local storage: this is recommended on diskless
platforms where there is no local storage attaching
to each processor.
• Optimization 3 (--predistribute-db), pre-distributing
database fragments to workers before the search be-
gins: especially useful in reducing data input time
when multiple database replicas need to be dis-
tributed to workers.
We have traced a large number of mpiBLAST ex-
perimental executions with the SST/macro simula-
tor to validate the simulator and predict the appli-
cation’s performance on a large-scale parallel ma-
chine. We executed our SST/macro simulation of the
mpiBLAST application on two different platforms:
a multi-core Linux machine and a distributed mem-
ory cluster system. The local machine consisted of a
2.66GHz Intel Core(TM)2 Duo CPU and 2GB mem-
ory. The cluster system is composed of 113 nodes,
where each node contains two 3.2GHz Intel Xeon
CPU and 2GB memory.
4.1 Validation of SST/macro with
mpiBLAST
SST/macro has been recently released (SST/Macro,
2011) and it has not yet been exposed to testing out-
side of its development environment at Sandia Na-
tional Labs. For this reason, in this section we briefly
mention our findings in our efforts to validate the
accuracy of the SST/macro simulator. The simula-
tor was validated with mpiBLAST results using de-
SIMULTECH 2011 - 1st International Conference on Simulation and Modeling Methodologies, Technologies and
Applications
70
0 10 20 30 40 50 60 70
0
200
400
600
800
1000
1200
1400
Number of Processors
Time (sec)
Local−Observed
Local−Simulated
Cluster−Observed
Cluster−Simulated
Figure 3: Comparison of observed and simulated runtimes
both on the local machine and the cluster system.
fault bandwidth (2.5 GB/s) and latency (1.3 µs) on
both testing environments. We used processor counts
from 8 to 64, and traces were collected using the
lightweight DUMPI library. Figure 3 shows the simu-
lated walltime versus the elapsed realtime for the sim-
ulation driven by these DUMPI traces.
We applied the concept of K-L divergence
(Emmert-Streib and Dehmer, 2008) to evaluate the
similarity between the results from observed and sim-
ulated run time across the tested systems. K-L diver-
gence is a non-commutativemeasure of the difference
between two samples P and Q typically P represent-
ing the “true” distribution and Q representing arbi-
trary distribution. Therefore we set P as simulation
results and Q as SST/macro DUMPI-driven runtimes
with varying total CPUs. The K-L divergence is de-
fined to be
D
KL
(P||Q) =
∑
i
P(i)log
P(i)
Q(i)
(1)
where Q(i) 6= 0. A smaller value of the K-L diver-
gence variable signifies greater similarity between the
two distributions.
Table 1 shows the K-L divergence distance. We
carefully analyzed the resulting K-L distance and
found out that the SST/macro trace clock times are
very close to real simulation wall-times on both the
local machine and the cluster system.
Table 1: The absolute distance and K-L divergence.
Method Local Cluster
K-L divergence 6.09 11.55
16 32 64
0
50
100
150
200
250
300
350
Number of Processors
Time (sec)
No Optimization
Optimization 1
Optimization 2
Optimization 3
Optimization 1+2+3
Figure 4: Scalability of 5 different optimization strategies
on a 113-node cluster system. Regular bars represent SST/-
macro DUMPI-driven simulation times with different opti-
mizations and the core bars represent observed time.
4.2 Simulation of mpiBLAST
Optimizations
To evaluate the various optimizations of mpiBLAST
that we mentionedearlier in this section, we run SST/-
macro and collected the simulation DUMPI traces
using 16, 32, and 64 processors on the cluster sys-
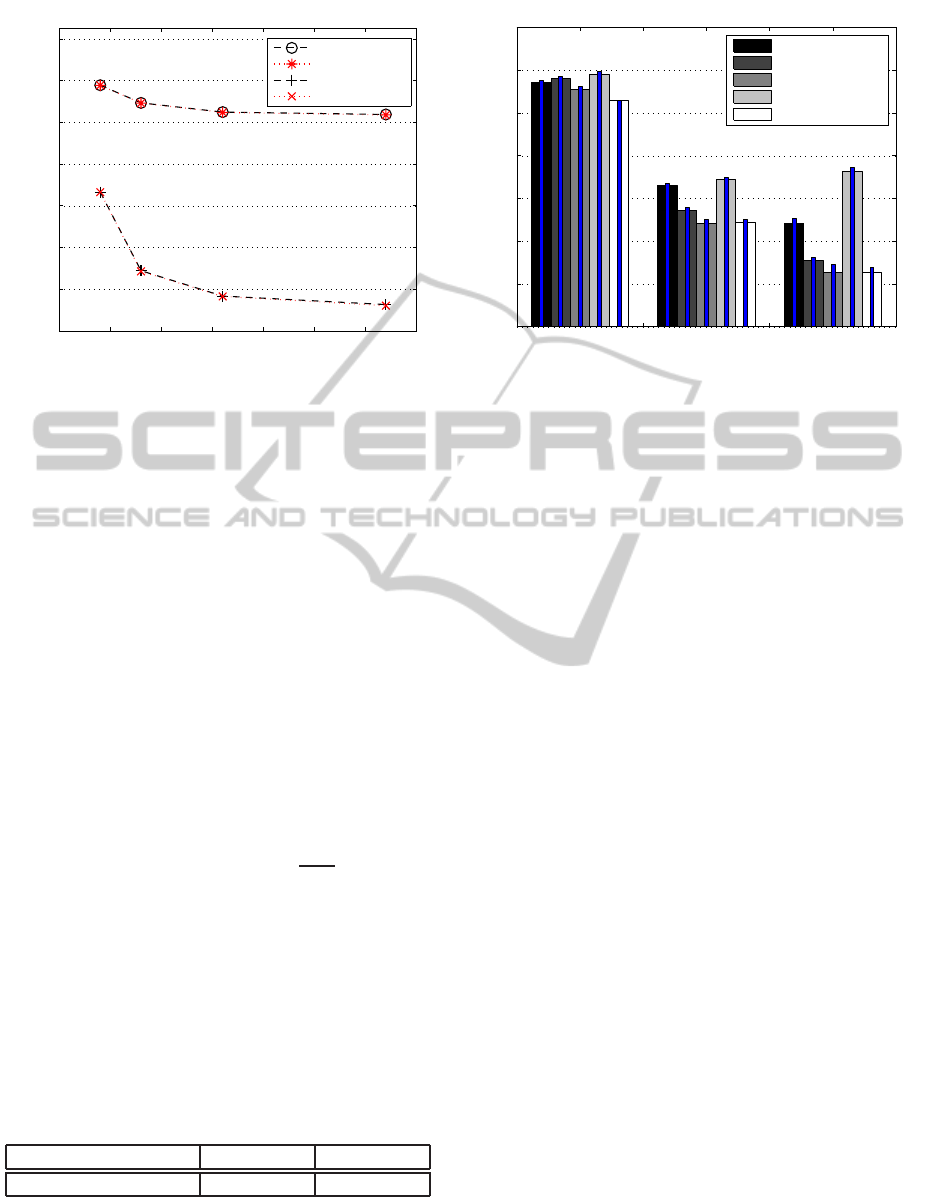
tem. Figure 4 shows the scalability and efficiency of
each approach. The y-axis shows the total execution
time in seconds for all processes of each approach,
and the x-axis represents the number of processors
that we used for our simulation runs. In our dia-
gram we use the following notations: Optimization 1
is the enabling of parallel output, Optimization 2 uses
virtual fragments, and Optimization 3 pre-distributes
database to workers before search begins. In addition,
we also tested a version that includes all three mpi-
BLAST refinements named Optimization 1+2+3, and
a version that excludes all optimizations named No
Optimizations. The simulation results indicate that for
our selected genome analysis, the sequence matching
of the Aedes aegypti genome using sequences of size
1MB, Optimization 2: the use of virtual fragments
provides the best scalability and efficiency. Optimiza-
tion 2 leads to a speed-up of a factor of 2 or more
compared to our No Optimization solution when exe-
cuted on 64 nodes of our cluster system. This finding
is not surprising given the exponential increase of the
cost of accessing global memory with the increase of
the participating compute nodes. Enabling the master
process to collect and write output (Optimization 1)
also led to a performance increase by about a factor
of 2 on our 64 node execution. Our tests indicated th-
EVALUATING PERFORMANCE OPTIMIZATIONS OF LARGE-SCALE GENOMIC SEQUENCE SEARCH
APPLICATIONS USING SST/MACRO
71

at in all execution scenarios the use of static work
pre-distribution alone (Optimization 3) led to a sig-
nificant overhead in our genome sequencing analy-
sis and slowed down the execution time. However,
when combined with Optimization 1 and Optimiza-
tion 2, static work pre-distribution did not lead to per-
formance loss and even helped increase the speed of
execution in certain scenarios. Enabling Optimization
2 helped in achieving faster execution in the tests we
performed using 16 and 32 nodes, however the ob-
served speed-up was not as significantly high as with
the scenario with 64 nodes. Intuitively, this result
demonstrates that Optimization 2 provides excellent
scalability, however, due to the overhead of comput-
ing the fragments, the methodology is effective only
when we have a system with a higher degree of par-
allelism. The graph in Figure 4 shows the same trend
for Optimization 1, where enabling parallel output
even deteriorated the execution time for the scenario
of using 16 cluster nodes.
5 CONCLUSIONS
The application of hardware/software co-design has
been a feature of embedded system designs for a long
time. So far, hardware/software co-design techniques
have found little application in the field of high-
performance computing. The multi-core paradigm
shift has left both software engineers and computer
architects with a lot of challenging dilemmas. The
application of hardware/software co-design for HPC
systems will allow for a bi-directional optimization of
design parameters where software specifications and
behavior drive hardware design decisions and hard-
ware constraints are better understood and accounted
for in the implementationof effectiveapplication soft-
ware. The use of cycle accurate simulation tools
provides the data and insights to estimate the per-
formance impact on an HPC applications when it is
subjected to certain architectural constraints. In this
work we demonstrated the application of a newly de-
veloped open-source cycle-accurate macroscale sim-
ulator (SST/macro) for the evaluation and optimiza-
tion of data intensive genome sequence matching al-
gorithms. We performed both trace-driven simula-
tion and simulation based on application modeling.
In our experimental set-up, we run an mpiBLAST
sequence matching algorithm using 1MB sequences
of the genome of the yellow fever mosquito, Aedes
aegypti. Using this data intensive application as
a canonical example, we validated the accuracy of
SST/macro. In addition, the analysis of our perfor-
mance data indicated that the use of dynamic data fr-
agmentation leads to significant performance gains
and high scalability on a distributed memory clus-
ter system. The framework we have presented in
this work allows for the evaluation and optimization
of mpiBLAST application on a wide variety of plat-
forms, ranging from a conventional workstation to a
system allowing levels of parallelism that are not ob-
tainable by existing supercomputers. This simulation
ability can play a crucial role for the effective de-
sign and implementation of large-scale data intensive
applications to be executed on the future multi-core
hardware platforms, that often could include a wide
variety of features including a heterogenous design of
CPUs, GPUs, and even FPGAs. In our future work,
we intend to further develop and distribute a full-scale
SST/macro model implementation of the entire mpi-
BLAST library and make it available as a part of the
SST/macro simulation distribution.
ACKNOWLEDGEMENTS
We express our gratitude to our team members of the
SST/macroscale simulator team at Sandia National
Laboratories, Livermore, CA: Scott Cranford, David
Evensky, Joe Kenny, Nicole Lemaster, Jackson Mayo,
and Ali Pinar. In addition, we thank Adrian Sandu
from Virginia Tech and Philip Bell from the Univer-
sity of Central Florida, and the anonymous referees
from SIMULTECH 2011 for providing helpful com-
ments and suggestions.
REFERENCES
Adve, V., Bagrodia, R., Deelman, E., and Sakellar-
iou, R. (2002). Compiler-optimized simulation of
large-scale applications on high performance architec-
tures. Journal of Parallel and Distributed Computing,
62(3):393–426.
Altschul, S., Gish, W., Miller, W., Myers, E., and Lipman,
D. (1990). Basic local alignment search tool. Journal
of Molecular Biology, 215:403–410.
Altschul, S., Madden, T., Sch¨affer, A., Zhang, J., Zhang,
Z., Miller, W., and Lipman, D. (1997). Gapped
BLAST and PSI-BLAST: a new generation of protein
database search programs. Nucleic Acids Research,
25(17):3389–3402.
Bjornson, R., Sherman, A., Weston, S., Willard, N., and
Wing, J. (2002). TurboBLAST(r): A Parallel Im-
plementation of BLAST Built on the TurboHub. In
Proc. International Parallel and Distributed Process-
ing Symposium (IPDPS’02), pages 183–190.
Braun, R., Pedretti, K., Casavant, T., Scheetz, T., Birkett,
C., and Roberts, C. (2001). Parallelization of Local
SIMULTECH 2011 - 1st International Conference on Simulation and Modeling Methodologies, Technologies and
Applications
72

BLAST Service on Workstation Clusters. Future Gen-
eration Computer Systems, 17(6):745–754.
Dally, W. and Towles, B. (2003). Principles and Practices
of Interconnection Networks. Morgan Kaufmann Pub-
lishers Inc., San Francisco, CA, USA.
Darling, A., Carey, L., and Feng, W. (2003). The Design,
Implementation, and Evaluation of mpiBLAST. In
Proceedings of ClusterWorld 2003.
Emmert-Streib, F. and Dehmer, M. (2008). Information
Theory and Statistical Learning. Springer Publishing
Company, Incorporated.
Gardner, M., Feng, W., Archuleta, J., Lin, H., and Ma, X.
(2006). Parallel Genomic Sequence-Searching on an
Ad-Hoc Grid: Experiences, Lessons learned, and Im-
plications. In IEEE/ACM International Conference
for High-Performance Computing, Networking, Stor-
age and Analysis (SC’06).
Janssen, C., Adalsteinsson, H., Cranford, S., Kenny, J.,
Pinar, A., Evensky, D., and Mayo, J. (2010). A Simu-
lator for Large-Scale Parallel Computer Architectures.
Inter. Jour. of Distributed Systems and Technologies,
1(2):57–73.
Kn¨upfer, A., R.B., Brunst, H., Mix, H., and Nagel, W.
(2006). Introducing the Open Trace Format (OTF).
In Alexandrov, V., van Albada, G., Sloot, P., and Don-
garra, J., editors, Int. Conf. on Computational Science,
volume 3992 of Lecture Notes in Computer Science,
pages 526–533. Springer.
Lin, H., Balaji, P., Poole, R., Sosa, C., Ma, X., and Feng, W.
(2008). Massively Parallel Genomic Sequence Search
on the Blue Gene/P Architecture. In Proc. ACM/IEEE
conference on Supercomputing (SC’08), pages 33:1–
33:11, Piscataway, NJ, USA. IEEE Press.
Lin, H., Ma, X., Chandramohan, P., Geist, A., and Sam-
atova, N. (2005). Efficient Data Access for Par-
allel BLAST. In Proc. International Parallel and
Distributed Processing Symposium (IPDPS’05), page
72.2, Washington, DC, USA. IEEE Computer Society.
Lin, H., Ma, X., Feng, W., and Samatova, N. (2011). Co-
ordinating Computation and I/O in Massively Parallel
Sequence Search. IEEE Transactions on Parallel and
Distributed Systems, 22(4):529–543.
Mathog, D. (2003). Parallel BLAST on split databases.
Bioinformatics, 19(14):1865–1866.
Mintchev, S. and Getov, V. (1997). PMPI: High-Level Mes-
sage Passing in Fortran 77 and C. In Proc. Inter. Con-
ference and Exhibition on High-Performance Com-
puting and Networking (HPCN Europe ’97), pages
603–614, London, UK. Springer-Verlag.
MPI (2009). MPI (Message Passing Interface) standards
documents, errata, and archives of the MPI Forum.
http://www.mpi-forum.org.
Prakash, S., Deelman, E., and Bagrodia, R. (2000).
Asynchronous Parallel Simulation of Parallel Pro-
grams. IEEE Transactions on Software Engineering,
26(5):385–400.
Riesen, R. (2006). A Hybrid MPI Simulator. In IEEE Inter.
Conf. on Cluster Computing 2006, pages 1–9.
Schuster, S. (2007). Next-generation sequencing transforms
today’s biology. Nature Methods, 5(1):16–18.
Sherwood, T., Perelman, E., and Hamerly, G. (2002). Auto-
matically Characterizing Large Scale Program Behav-
ior. In 10th International Conference on Architectural
Support for Programming Languages and Operating
Systems (ASPLOS 2002), pages 45–57.
SST/Macro (2011). SST: The Structural Sim-
ulation Toolkit, SST/macro the Macroscale
Components, Open Source Release.
http://sst.sandia.gov/about sstmacro.html.
Stroustrup, B. (2000). The C++ Programming Lan-
guage. Addison-Wesley Longman Publishing Co.,
Inc., Boston, MA, USA.
Susukita, R., Ando, H., Aoyagi, M., Honda, H., Inadomi,
Y., Inoue, K., Ishizuki, S., Kimura, Y., Komatsu, H.,
Kurokawa, M., Murakami, K. J., Shibamura, H., Ya-
mamura, S., and Yu, Y. (2008). Performance predic-
tion of large-scale parallell system and application us-
ing macro-level simulation. In Proc. ACM/IEEE con-
ference on Supercomputing SC ’08, pages 20:1–20:9,
Piscataway, NJ, USA. IEEE Press.
Thorsen, O., Smith, B., Sosa, C., Jiang, K., Lin, H., Peters,
A., and Feng, W. (2007). Parallel genomic sequence-
search on a massively parallel system. In Proc. Int.
Conf. on Computing Frontiers (CF ’07), pages 59–68,
New York, NY, USA. ACM.
Top500 (2011). Top 500 SuperComputers Ranking at
March 2011. http://www.top500.org.
Underwood, K. D., Levenhagen, M., and Rodrigues, A.
(2007). Simulating Red Storm: Challenges and Suc-
cesses in Building a System Simulation. In Proc. In-
ternational Parallel and Distributed Processing Sym-
posium (IPDPS’07), pages 1–10, Los Alamitos, CA,
USA. IEEE Computer Society.
Vectorbase (2010). NIAID Bioinformatics Resource Cen-
ter for Invertebrate Vectors of Human Pathogens.
http://www.vectorbase.org.
Zheng, G., Wilmarth, T., Jagadishprasad, P., and Kal´e, L.
(2005). Simulation-based performance prediction for
large parallel machines. Int. Jour. Parallel Program.,
33(2):183–207.
EVALUATING PERFORMANCE OPTIMIZATIONS OF LARGE-SCALE GENOMIC SEQUENCE SEARCH
APPLICATIONS USING SST/MACRO
73
