
TOWARD A NON INVASIVE CONTROL OF APPLICATIONS
A Biomedical Approach to Failure Prediction
Ilenia Fronza, Alberto Sillitti, Giancarlo Succi and Jelena Vlasenko
Free University of Bolzano, Bozen, Piazza Domenicani, Domenikanerplatz, 3, I-39100 Bolzano – Bozen, Italy
Keywords: Failure prediction, Survival analysis, Cox PH model, Event sequence data, Log files.
Abstract: Developing software without failures is indeed important. Still, it is also important to detect as soon as
possible when a running application is likely to fail, so that corrective actions can be taken. Following the
guidelines of Agile Methods, the goal of our research is to develop a statistical prediction model for failures
that does not require any additional effort on the side of the developers of an application; the key concept is
that the developers concentrate on the code and we use the information that is naturally generated by the
running application to assess whether an application is likely to fail. So the developers concentrate only on
providing direct value to the customer and then the model takes care of informing the environment of the
possible crash. The proposed model uses as input data that is commonly produced by developers: the log
files. The statistical prediction model employed comes from biomedical studies about cancer survival
prediction based on gene expression profiles where gene expression measurements and survival times of
previous patients are used to predict future patients' survival. One of the most prominent models is the Cox
Proportional Hazards (PH) model. In this work, we draw a parallel between our context and the biomedical
one; we consider types of operations as genes, and operations and their multiplicity in the sequence as
expression profiles. Then, we identify signature operations applying the above mentioned Cox PH model.
We perform a prototypical analysis using real-world data to assess the suitability of our approach. We
estimate the confidence interval of our results using Bootstrap.
1 INTRODUCTION
Moving from proactive maintenance to preventive
maintenance may reduce wastes caused by
downtime costs. Therefore, much of current research
is focused on predicting software failures with a
sufficient lead of time before actual failure.
Failure prediction models are usually built upon
historic data to predict the occurrence of failures. In
general, some predictor variables are collected or
estimated at the beginning of a project and fed into a
model. This approach is reasonable for traditional
processes.
One of the key guidelines of Agile Methods is to
consider feedback, since "it is rare to find control
without feedback, because feedback gives much
better control and predictability than attempting to
control complicated processes with predefined
algorithms" (Poppendieck and Poppendieck, 2003).
Therefore, an Agile failure prediction model is
required to use feedback to refine itself during the
development process.
Moreover, to cope and even accomodate Agile
Methods, a failure prediction model needs not to
require any additional effort on the side of the
developers of an application. Developers need to
eliminate waste of time and to concentrate on the
code providing direct value to the customer.
Therefore, failure prediction models are required to
inform of the possible crash using information that is
naturally generated by the running application.
Being cognizant of the mentioned challenges, the
key objective of our study can be outlined as
follows: we propose a new, incremental failure
prediction tool which uses data from previous
iterations to refine itself at every iteration.
Our idea is to use as input for the model data that
is commonly produced by developers: the log files.
These files contain a series of events marked with
their occurrence times. Thus, we deal with
prediction of failure event(s) based on the analysis
of event sequence data.
To this purpose, some techniques already exist
and can be classified into design-based methods and
data-driven rule-based methods. In a design-based
83
Fronza I., Sillitti A., Succi G. and Vlasenko J..
TOWARD A NON INVASIVE CONTROL OF APPLICATIONS - A Biomedical Approach to Failure Prediction.
DOI: 10.5220/0003493000830091
In Proceedings of the 13th International Conference on Enterprise Information Systems (ICEIS-2011), pages 83-91
ISBN: 978-989-8425-54-6
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

method, the expected event sequence is obtained
from the system design and is compared with the
observed event sequence. A system logic failure can
be identified by use of this comparison. Untimed and
timed automata models, and untimed and timed Petri
net models have been proposed in (Chen et al., 2009,
Sampath et al., 1994, Srinivasan and Jafari, 1993);
time template models (Holloway 1996, Pandalai and
Holloway, 2000) establish when events are expected
to occur basing on timing and sequencing
relationships of events, which are generated from
either timed automata models (system design) or
observations of manufacturing systems. The major
disadvantage of these methods is that in many cases,
events occur randomly and thus there is no system
logic design information available.
Data-driven rule based methods do not require
system logic design information. These methods are
made of two phases: 1) identification of the temporal
patterns, i.e., the sequences of events that frequently
occur, and then development of prediction rules
based on these patterns (Agrawal 1996, Mannila et
al., 1997), and 2) prediction of the occurrence of a
failure event basing on the time relationships among
events (Dunham, 2003; Klemettinen 1999; Li et al.,
2007).
Survival analysis models have been also
proposed to solve this issues. Code metrics are given
as input to Cox PH model in (Wedel et al., 2008) to
analyze failure time data from bug reports of open
source software. In (Li et al., 2007) the Cox PH
model is used to predict system failures based on the
time-to-failure data extracted from the event
sequences. Only event combinations identified as
signatures are treated as explanatory variables in the
Cox PH model.
We propose a mechanism for lean, non invasive
control of applications. The idea is to build devices
that read logs of running applications and signal the
likely crash of such systems, thus adding no "muda"
(Liker 2003) to the development of each application.
We propose to collect (Dubson 2006) data using
Automated In-process Software Engineering
Measurement and Analysis (AISEMA) systems
(Coman et al., 2009). Then, we address the analyse
and decide phases (Dubson 2006) looking at
biological studies about cancer survival prediction
based on gene expression profiles. The idea is to
follow biomedical guidelines to:
analyse data and identify signature operations of
failure;
assign a risk score to future sequences of
operations;
decide if these sequences will fail or not.
This mechanism looks at the running system as a
“black box”, meaning that we do not have any other
information about the system except the log files.
Altogether, the proposed approach is:
lean, because there is no need of an explicit
mechanism on the applications (Liker, 2003);
non-invasive, because data is collected by an
AISEMA system (Coman et al., 2009);
intended to become an incremental failure
prediction tool which is built after each end of a
sequence of operations and uses data from previous
iterations to refine itself at every iteration.
In this work, we first draw a parallel between our
context and the biomedical one, since they deal with
different types of entities. As a result of this parallel,
we consider types of operations as genes, and
operations and their multiplicity in the sequence as
expression profiles. Signature operations of failures
are identified applying the Cox PH model, one of the
most prominent models in biomedical studies.
Then, we present a prototypical analysis using
real-world data, consisting in log files collected
during approximately 3 months of work in an
important Italian company that prefers to remain
anonymous. We propose to use Bootstrap methods
to determine the confidence interval of our results
The paper is structured as follows: in Section 2,
we present some background about survival analysis
together with the Cox Proportional Hazard (PH)
model and its applications; in Section 3, we
introduce our approach to failure prediction. In
Section 4, sample applications are presented. In
Section 5 we discuss our results.
2 BACKGROUND
2.1 Survival Analysis
The term “survival data” is used in a broad sense for
data involving time to the occurrence of a certain
event. The event of interest is usually death, the
appearance of a tumor, the development of some
disease, or some negative experience; thus, the event
is called “failure”. However, survival time may be a
positive event, like time to return to work after an
elective surgical procedure (Kleinbaum and Klein,
2005), or cessation of smoking, and so forth.
In the past decades, applications of the statistical
methods for survival data analysis have been
extended beyond biomedical research to other fields,
for example criminology (Benda, 2005; Schmidt and
Witte, 1989), sociology (Agerbo, 2007; Sherkat and
ICEIS 2011 - 13th International Conference on Enterprise Information Systems
84

Ellison, 2007), and marketing (Barros and Machado,
2010; Chen et al., 2009).
The study of survival data has previously
focused on predicting the probability of response,
survival, or mean lifetime, and comparing the
survival distributions of experimental animals or of
human patients.
In recent years, the identification of risk and/or
prognostic factors related to response, survival, and
the development of a disease has become equally
important (Persson, 2002).
The analysis of survival data is complicated by
issues of censoring. Censored data arises when an
individual’s life length only is known to occur in a
certain period of time (Kleinbaum and Klein, 2005).
2.2 The Cox Proportional Hazards
(PH) Model
Introduced by D.R. Cox in 1972 (Cox, 1972), the
semiparametric Cox PH model estimates the hazard
function, which assesses the instantaneous risk of
failure.
The data, based on a sample of size n, consists of
(x
i
, t
i
, δ
i
), i = 1, ... , n where:
x
i
is the vector of p covariates or risk factors for
individual i which may affect the survival
distribution of the time to event;
t
i
is the time under observation for individual i;
δ
i
is the event indicator (δ
i
=1 if the event has
occurred, δ
i
=0 otherwise).
The Cox PH model has became the most commonly
used model to give an expression for the hazard at
time t with a given specification of p covariates x:
(1)
The Cox PH model formula says that the hazard at
time t is the product of two quantities:
the baseline hazard function h
0
(t), which is equal
for all the individuals. It may be considered as a
starting version of the hazard function, prior to
considering any of the x´s;
the exponential expression e to the linear sum of
β
i
x
i
, where the sum is over the p explanatory x
variables.
The Cox PH model focuses on the estimation of the
vector of regression coefficients β leaving the
baseline hazard unspecified. In the p < n setting
(Bøvelstad, 2010), β´s are estimated by maximizing
the log partial likelihood, which is given by:
(2)
The key assumption of the Cox PH model is
proportional hazards; this assumption means that the
hazard ratio (defined as the hazard for one individual
over the hazard for a different individual) is constant
over time.
This model is widely used because of its
characteristics:
even without specifying h
0
(t), it is possible to
find the β´s;
no particular form of probability distribution is
assumed for the survival times. A parametric
survival model is one in which survival time (the
outcome) is assumed to follow a known distribution.
The Cox PH model is not a fully parametric model.
Rather it is a semi-parametric model because even if
the β's are known, the distribution of the outcome
remains unknown;
it uses more information than the logistic model,
which considers a (0,1) outcome and ignores
survival times and censoring. Therefore it is
preferred over the logistic model when survival time
is available and there is censoring (Kleinbaum and
Klein, 2005).
2.3 Applications of the Cox PH Model
The Cox PH model was first used in biomedical
applications (Collett, 1994), mainly to determine
prognostic factors that affect the survival time of
patients to some type of diseases (mostly cancers)
(Eliassen et al., 2010, Pope et al., 2002).
After the completion of DNA sequencing, the
Cox PH model has been applied to gene expression
data to detect novel subtypes of a disease, or to
explore the potential associations between the high-
dimensional gene expression data and some clinical
outcome.
Cancer patient survival based on gene expression
profiles is an important application of genome-wide
expression data. The goal is to identify the optimal
combination of the gene expression data in
predicting the risk of cancer death.
The most common approach to model covariate
effects on cancer survival is the Cox PH model,
which takes into account the effect of censored
observations (Bøvelstad et al., 2007, Hao et al.,
2009, Yanaihara et al., 2006, Yu et al., 2008). Gene
expression measurements and survival times of
previous patients are used to obtain a prediction rule
that predicts the survival of future patients.
TOWARD A NON INVASIVE CONTROL OF APPLICATIONS - A Biomedical Approach to Failure Prediction
85

Given n patients diagnosed with a specific type
of cancer, for each of the n patients the observations
(x
i
, t
i
, δ
i
) are available, where:
x
i
= (x
i1
, … , x
ip
) contains the measurements of p
gene expression values for patient i; i = 1, ... , n;
t
i
is the time under observation of patient i;
the censoring index δ
i
keeps track of whether
patient i died (δ
i
= 1) or was censored (δ
i
= 0), i.e.,
patient i was still alive at the end of the study or if
she or he dropped out of the study or died from
another cause.
Applications of the Cox PH model can be found in
several fields of research other than biomedicine,
such as criminology (Benda, 2005; Schmidt and
Witte, 1989), sociology (Agerbo, 2007; Sherkat and
Ellison, 2007), marketing (Barros and Machado,
2010; Chen et al., 2009), and cybernetic (Li et al.,
2007). There is also an application of the Cox PH
model to software data in (Wendel 2008) where,
using as input code metrics, failure time data coming
from bug reports have been analysed.
2.4 The Bootstrap Method
In statistics the average is often used to guess the
overall behavior of a population using the
information coming from a sample; depending on
the structure of the population, the average could
then be implemented by taking the means, the
median, or the mode (Fisher and Hall, 1991).
However, it is interesting also to know how reliable
is an estimate done with the average. To this end,
other statistical properties are often used, based on
the structure of the population (e.g., standard
deviations, percentiles, histograms). Clearly, a
reliable estimation of the confidence interval can
provide a very valuable information to the user of an
estimation.
To this end the Bootstrap technique has been
devised (Efron, 1987). Bootstrap is a computer-
based (it can be both parametric and non-parametric)
method to compute estimated standard errors,
confidence intervals, and hypothesis testing (Efron
and Tibshirani, 1993).
The Bootstrap algorithm first creates n datasets
of the same size of the original sample by randomly
picking elements of it with replacement; this
technique is called resampling. The number of the
generated sets needs to be high; a suitable number
could be 1000.
Then, the Bootstrap algorithm computes the
averages of each of the newly generated datasets –
the specific kind of average depends from the
statistical properties of the population. The results
are then used to determine the confidence interval of
the population from which the original sample was
drawn.
In this study we want to determine the reliability
of our estimations for failures. However, we have a
very limited dataset and we do not know the
parameters of the random variables of interest, and
the overall transformations are non-linear.
Therefore, we propose to use Bootstrap method.
Each time an estimation of failure or success is
done, we use Bootstrap to determine the reliability
of it, that is, the percentage of times in which the
results remain consistent with the original
prediction.
3 APPROACH
3.1 Schema of the Approach
To build devices that read logs of running
applications and signal the likely crash of such
systems, we have to address the four main activities
reported in (Dobson et al., 2006): 1) collect, 2)
analyse, 3) decide, and 4) act.
Figure 1 presents a schematic view of the
proposed approach.
While the system is running, an AISEMA system
(Coman et al., 2009) collects data to track the actual
execution path. In this work, we look at the running
system as a “black box”, meaning that we do not
have any other information about the system except
the log files.
The monitoring process takes log data as input,
analyses them, and basing on the analysis performed
decides the “likely failure” of the running
application and gives to the supervisor a message.
Figure 1: Schema of the devices that read logs of a
running system and signal the likely failure.
The supervisor can act directly on the running
system to avoid the predicted failure, or send an alert
to the outside world. Possible actions could be to
ICEIS 2011 - 13th International Conference on Enterprise Information Systems
86

abort the running system, to restart it, to dynamically
load components, or to inform the running system if
it was a suitably structured autonomic system
(Müller et al., 2009).
The monitoring process is based on the Cox PH
model. Our context seems to have the characteristics
needed to apply Cox PH model; in fact, survival
time and censored observations are available.
Moreover, the Cox PH model needs no assumption
of a parametric distribution for the event sequence
data. Therefore, we can benefit of the following
advantages:
we can discover information that may be hidden
by the assumption of a specific distribution (Yu et
al., 2008);
we obtain results comparable to the parametric
model even without this assumption (Kalbfleisch
and Prentice, 2002).
Our context is completely different from the
biomedical one. Therefore, before applying the Cox
PH model, we first drew a parallel between the two
contexts to provide an interpretation of the
biomedical entities in our case.
3.2 Parallel between Our Context and
the Biomedical One
The idea behind biomedical studies is that different
gene expression profiles may result in different
survival times. In software processes, the activities
performed may affect the time of survival to a
failure. Therefore, in our context, types of operations
in some sense play the role of genes.
The expression profile may be formed by each
operation's multiplicity, since it provides
information about the level of presence of the
operation.
Time under observation of a sequence is defined
as its length in terms of time, meaning that we
evaluate it as the difference between the time stamp
of the last operation and the time stamp of the first
operation in the sequence.
In our case, the “observed event” is the end of
the sequence because of a failure. Therefore, we
have the following definition: δ
i
=1 if i is a failure, δ
i
=0 otherwise.
Our type of censoring is type II with a
percentage of 100% (Lee and Wang, 2003), meaning
that δ
i
is never equal to zero because of the end of
the period of observation.
Table 1 summarizes the interpretation of
biological entities in our context. In biomedical
studies, gene expression measurements and survival
times of previous patients are used to obtain a
prediction rule that predicts the survival of future
patients. In this work, we consider types of
operations as genes, and operations and their
multiplicity in the sequence as expression profiles.
Table 1: Parallel between our context and the biomedical
one.
Variable Biological data Our data
t
i
Time under
observation of
patient i
Time under
observation of
sequence i (last
date – first date)
δ
i
δ
i
=1 if survival
time is observed
δ
i
=0 elsewhere
δ
i
=1 for failures
δ
i
=0 elsewhere
x
i
=
(x
i1
, …, x
ip
)
x
ij
is the
expression value
of gene j in
patient i
x
ij
is the
multiplicity of
operation j in
sequence i
i=1, …, n j=1, …, p
3.3 Structure of the Monitoring
Process
3.3.1 Dimensional Reduction of the Problem
Our method consists of the following steps to get
from raw logs to temporal event sequences (Zheng
et al., 2009) to be used as input for the analyses:
data are parsed to extract operations together
with their associated time stamps and severities for
each event in the log file;
duplicate rows are deleted together with logs that
are missing information in one or several of the
fields Operation, Time stamp, Severity;
sequences of activities are extracted: a new
sequence starts either if there is a “Log in” operation
or if the day changes.
Failures are defined as sequences containing at least
one severity “Error”. Table 2 summarizes the
definitions used in this work.
Table 2: Definitions used in this work.
Notion Definition
sequence A chronologically ordered set of log entries
in the maximum time frame of 1 day. Two
sequences during the same day are
separated by a “Log in” operation
Failure A sequence containing at least one severity
“Error”
TOWARD A NON INVASIVE CONTROL OF APPLICATIONS - A Biomedical Approach to Failure Prediction
87

3.3.2 Preparation of the Input for the Cox
PH Model
Each temporal event sequence i is described by (x
i
,
t
i
, δ
i
), where:
x
i
= (x
i1
, ..., x
ip
), and x
ij
is the multiplicity of
operation j in sequence i;
t
i
is the lifetime of the sequence, defined as the
difference between the last time stamp and the first
time stamp of sequence i;
δ
i
= 1 when the event is “observed” (failure
sequences) and δi = 0 elsewhere (censored
observations).
3.3.3 Training of the Model
We propose to follow the guidelines given in
biomedical studies (Bøvelstad et al., 2007, Hao et
al., 2009, Yanaihara et al., 2006, Yu et al., 2008) to
analyse data about past sequences and decide about
the likely failure of the current sequence of
operations of the running system. In particular, the
training of the monitoring process includes the
following steps:
the Schoenfeld test (Hosmer et al., 2008;
Kalbfleisch and Prentice, 2002; Kleinbaum and
Klein, 2005) is applied to select the operations
satisfying the PH assumption;
hazard ratios from a multivariate Cox PH model
are used to identify which operations are associated
to a failing end of sequences. The k operation
included in the model are those operations whose
multiplicity in the sequence is expected to be related
to failure. Protective operations are defined as those
with hazard ratio for failure lower than 1. High-risk
operations are defined as those with hazard ratio for
failure greater than one;
the k-operations signature risk score RS is
assigned to each sequence in the dataset, according
to the exponential value of a linear combination of
the multiplicity of the operation, weighted by the
regression coefficients derived from the
aforementioned Cox PH model;
the following values are extracted: 1) m, the third
quartile of risk scores of non failure sequences, and
2) M, maximum risk score of non failure sequences.
3.3.4 Deciding the “Likely Failure” and the
Associated Confidence Interval
The k-operations signature risk score RS is evaluated
for the new sequence, and basing on its value the
sequence is defined as:
“likely no failure” if RS ≤ m;
“likely failure” if RS ≥ M;
“still unknown” if m < RS < M.
In order to compute estimates of confidence
intervals of our dataset we propose to use the
Bootstrap method of resampling. We perform a
Bootstrap of 1000 times on the original data and
then we consider for each sequence the resulting
prediction.
The sets of the predictions is then used to
determine the likelihood of correctness of the
original prediction.
4 SAMPLE APPLICATIONS
To assess the suitability of our approach, we have
performed two prototypical analysis intended to
determine if it is worth pursuing the approach
further. We use two datasets consisting of log files
collected during approximately 3 months of work in
an important Italian company that prefers to remain
anonymous.
We prepared each dataset as discussed in Section
3.3.1 and Section 3.3.2. Then we assigned randomly
the sequences to training set (60%) or test set (40%).
Afterwards, we proceeded as discussed in Section
3.3.3 and Section 3.3.4 to 1) identify the k signature
operations of failure in the training set, 2) compute
the values of m and M, 3) assign the the k-operations
signature risk score to each sequence in the test set,
and 4) decide the “likely failure” of the sequences in
the test set.
4.1 First Sample
Table 3 contains the training set pre-processing
summary of the first sample
Table 3: Training set pre-processing summary (first
sample).
Notion Definition N %
Cases
available in
the analysis
Event
1
28 12.9
Censored 157 72.0
Total 185 84.9
Cases
dropped
Censored before the first
event in the stratum
33 15.1
Total 218 100
1
Dependent variable: survival time
Six out of the eight initial operations were
satisfying the proportional hazards assumption and
were therefore kept in the input dataset for the Cox
PH model. Three out of the six initial operations
ICEIS 2011 - 13th International Conference on Enterprise Information Systems
88
were kept in the output of the model. Table 4
contains the output of this model, together with the
definition of each operation as “protective” (or
“high-risk”) according to the exponential value of
the regression coefficient β.
Table 4: Output of Cox PH model on training set (first
sample).
β Sig Exp(β) definition
Operation 1 -0.40 0.013 0.961 Protective
Operation 2 0.06 0.015 1.006 High-risk
Operation 3 -0.52 0.055 0.592 protective
-2 Log Likelihood: 200.112
The comparison between failures and non
failures shows that higher risk scores have been
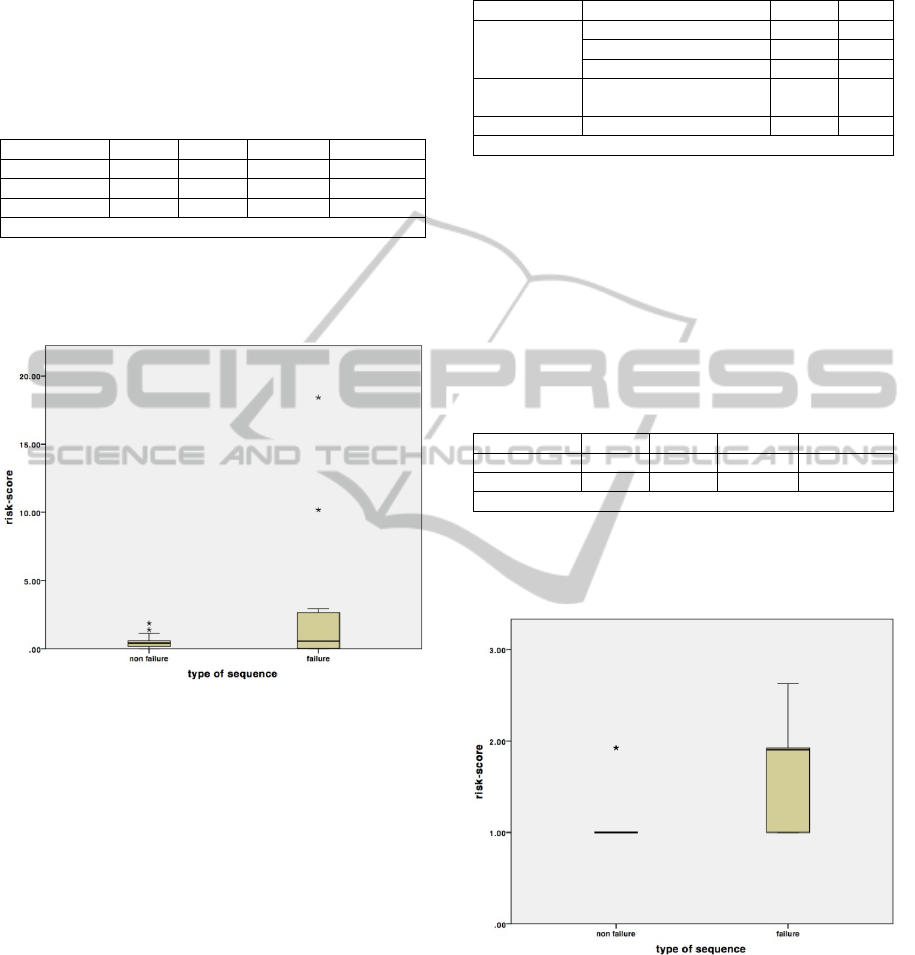
assigned to failure sequences (Figure 2).
Figure 2: Risk scores in failure and non-failure sequences
(first sample).
Altogether we obtained a value of m = 0.59 and
M = 1.85.
In the test set, the comparison of the risk scores
with m and M provides the following results: in 40%
of the cases the system is able to predict correctly
the failure and only in 1% of the cases a predicted
failure is not a failure, which means that a message
of expected failure is quite reliable. On the contrary,
the prediction of non-failure is not as reliable. 50%
of the failing sequences are predicted as non-failing.
4.2 Second Sample
Table 6 contains the training set pre-processing
summary of the second sample.
Table 5: Training set pre-processing summary (second
sample).
Notion Definition N %
Cases
available in
the analysis
Event
1
139 1.8
Censored 7445 98.2
Total 7584 100
Cases
dropped
Censored before the first
event in the stratum
0 0
Total 7584 100
1
Dependent variable: survival time
Six out of the ten initial operations were
satisfying the proportional hazards assumption and
were therefore given as input to the Cox PH model.
Two out of the six initial operations were kept in the
model. Table 7 contains the output of this model,
together with the definition of each operations as
“protective” (or “high-risk”) according to the
exponential value of the regression coefficient β.
Table 6: Output of the Cox PH model on the training set
(second sample).
β Sig Exp(β) definition
Operation 1 0.66 0.001 1.925 High-risk
Operation 2 0.32 0.001 1.380 High-risk
-2 Log Likelihood: 1746.471
The comparison between failures and non
failures shows that higher risk scores have been
assigned to failure sequences (Figure 3).
Figure 3: Risk scores in failure and non failure sequences
(second sample).
In this case, we obtain a value of m = 1.00 and M
= 1.93.
In the test set, the comparison of the risk scores
with m and M provides the following results: in
100% of the cases the approach taken is able to
predict correctly the failure, and in 99.6% of the
cases the system is able to predict correctly the non
TOWARD A NON INVASIVE CONTROL OF APPLICATIONS - A Biomedical Approach to Failure Prediction
89

failure. Both the messages of expected failure and
non failure appear to be quite reliable.
5 CONCLUSIONS
In this work we propose a mechanism for lean, non
invasive control of applications. We use an
AISEMA system (Coman et al., 2009) to collect
data; then we apply the Cox PH Model following the
biomedical guidelines to find the k signature
operations of failure and to decide about the “likely
failure” of future sequences
.
Altogether, the proposed approach presents the
following advantages:
it is lean, because there is no need for an explicit
mechanism on the application;
it is non invasive, because data is collected using
AISEMA systems;
data collection is low cost;
measurement is continuous and accurate;
developers' efficiency is not affected by data
collection since they can concentrate on their tasks
as usual.
The proposed mechanism is intended to become an
incremental failure prediction tool which is built
after each end of a sequence of operations and
refines itself at every iteration using data from
previous iterations.
Results from our preliminary, prototypical
analysis appear quite interesting and worth further
investigations; higher risk scores are assigned to
failure sequences in the test set. Overall, the
proposed approach appears very effective in the
identification of True Positives and True Negatives,
meaning that the sequences identified as leading to a
failure are very likely to lead to a failure.
Additional work is now needed to use Bootstrap
methods to determine the confidence interval of our
results; we are now accomplishing this task.
Moreover, we need to study more in-depth our
promising mechanism to determine the
generalizability of our results. To this end, we are
going to study more in-depth our model, trying to
generalize our results. In particular, we plan to
replicate the analysis on more industrial datasets.
We are also considering the following
approaches to achieve higher levels of precisions: 1)
the Cox PH model with strata to analyse covariates
not satisfying the PH assumption, 2) specific
techniques to manage datasets with a limited number
of cases (Bøvelstad, 2010), and 3) application of
Bootstrap methods in order to identify the
confidence interval of the prediction.
We should also investigate how other “black-
box” properties of applications (e.g., memory usage,
number of open files, processor usage) can be
considered to predict failures.
We will also deal with the bias introduced when
calculating survival time without considering the
duration of the last operation.
We will then try to predict a failure analysing
only an initial portion of a sequence, to obtain an
early estimation of failure, providing additional time
to take corrective actions.
Finally, the proposed model could be particularly
useful dealing with autonomic systems (Müller et
al., 2009).
ACKNOWLEDGEMENTS
We thank the Autonomous Province of South Tyrol
for supporting us in this research.
REFERENCES
Agerbo, E. 2007. High Income, Employment,
Postgraduate Education, and Marriage: a Suicidal
Cocktail among Psychiatric Patients. Archives of
General Psychiatry, 64, 12, 1377-1384.
Barros, C. P. and Machado, L. P. 2010. The Length of
Stay in Tourism. Annals of Tourism Research, 37, 3,
692-706.
Benda, B. 2005. Gender Differences in Life-Course
Theory of Recidivism: A Survival Analysis.
International Journal of Offender Therapy and
Comparative Criminology, 49, 3, 325-342.
Bøvelstad, H. M., Nygård, S., Størvold, H. L., Aldrin, M.,
Borgan, Ø., Frigessi, A., and Lingjærde, O. C. 2007.
Predicting Survival from Microarray Data: a
Comparative Study. Bioinformatics, 23, 16, 2080-
2087.
Bøvelstad, H. M. 2010. Survival Prediction from High-
Dimensional Genomic Data. Doctoral Thesis.
University of Oslo.
Chen,Y., Zhang, H., and Zhu, P. 2009. Study of Customer
Lifetime Value Model Based on Survival-Analysis
Methods. In World Congress on Computer Science
and Information Engineering (Los Angeles, USA,
March 31 – April 02, 2009), 266-270.
Coman, I. D., Sillitti, A., and Succi, G. 2009. A case-study
on using an Automated In-process Software
Engineering Measurement and Analysis system in an
industrial environment. In ICSE’09: International
Conference on Software Engineering (Vancouver,
Canada, May 16-24 May, 2009), pp. 89-99.
ICEIS 2011 - 13th International Conference on Enterprise Information Systems
90

Cox, D.R. 1972. Regression models and life-tables.
Journal of the Royal Statistical Society Series B, 34,
187-220.
Collett, D. 1994. Model ling Survival Data in Medical
Research. Chapman&Hall, London.
Dobson, S., Denazis, S., Fernández, A., Gaiti, D.,
Gelenbe, E., Massacci, F., Nixon, P., Saffre, F.,
Schmidt, N., and Zambonelli, F. 2006. A Survey of
Autonomic Communications. ACM Transactions on
Autonomous and Adaptive Systems, 1, 2, 223-259.
Efron, B. 1987. Better Bootstrap Confidence Interval.
Journal of the American Statistical Association, 82,
171-200.
Efron, B. and Tibshirani, R.: An Introduction to the
Bootstrap. Chapman & Hall (1993).
Eliassen, A. H., Hankinson, S. E, Rosner, B., Holmes, M.
D., and Willett, W. C. 2010. Physical Activity and
Risk of Breast Cancer Among Postmenopausal
Women. Archives of Internal Medicine, 170, 19, 1758-
1764.
Fisher, N. I. and Hall, P. 1991. Bootstrap algorithms for
small samples. Journal of Statistical Planning and
Inference, 27, 157-169.
Hao, K., Luk, J. M., Lee, N. P. Y., Mao, M., Zhang, C.,
Ferguson, M. D., Lamb, J., Dai, H., Ng, I. O., Sham,
P.C., and Poon, R.T.P. 2009. Predicting prognosis in
hepatocellular carcinoma after curative surgery with
common clinicopathologic parameters. BMC Cancer,
9, 398-400.
Hosmer, D. W., Lemeshow, S., and May, S. 2008. Applied
survival analysis: Regression modeling of time to
event data. Wiley.
Kalbfleisch, J. D. and Prentice, R. L. 2002. The statistical
analysis of failure time data. Wiley.
Kleinbaum, D. G. and Klein, M. 2005. Survival analysis:
a self-learning test (Statistics for Biology and Health).
Springer.
Lee E. T. and Wang, J. W. 2003. Statistical methods for
survival data analysis. Wiley.
Li, Z., Zhou, S., Choubey, S., and Sievenpiper, C. 2007.
Failure event prediction using the Cox proportional
hazard model driven by frequent failure sequences.
IEE Transactions, 39, 3, 303-315.
Liker, J. 2003. The Toyota Way. 14 Management
Principles from the World's Greatest Manufacturer.
McGraw Hill.
Mannila, H., Toinoven, H., and Verkamo, A. I. 1997.
Discovery of frequent episodes in event sequences.
Data Mining and Knowledge Discovery, 1, 259-289.
Müller, H. A., Kienle, H. M., and Stege, U. 2009.
Autonomic Computing: Now You See It, Now You
Don’t—Design and Evolution of Autonomic Software
Systems. In: De Lucia, A., Ferrucci, F. (eds.):
Software Engineering International Summer School
Lectures: University of Salerno 2009. LNCS, vol.
5413, pp. 32-54. Springer-Verlag.
Pandalai, D. N. and Holloway, L. E. 2000. Template
languages for fault monitoring of timed discrete event
processes. IEEE Transactions on Automatic Control
,
45, 5, 868-882.
Persson, I. 2002. Essays on the Assumptions of
Proportional Hazards in Cox Regression. Doctoral
Thesis. Uppsala University.
Pope, C. A., Burnett, R. T., Thun, M. J., Calle, E. E.,
Krewski, D., Ito, K., and Thurston, G. D. 2002. Lung
Cancer, Cardiopulmonary Mortality, and Long-term
Exposure to Fine Particulate Air Pollution. Journal of
the American Medical Association, 27, 9, 1132- 1141.
Poppendieck, M. and Poppendieck, T. 2003. Lean
software development: an Agile toolkit. Addison
Wesley.
Sampath, M., Sengupta, R., and Lafortune, S. 1994.
Diagnosability of discrete event systems. In:
International Conference on Analysis and
Optimization of Systems Discrete Event Systems
(Sophia, Antipolis, June 15 – 17, 1994), pp. 73-79.
Schmidt, P. and Witte, A. D. 1989. Predicting Criminal
Recidivism Using “Split Population” Survival Time
Models. Journal of Econometrics, 40, 1, 141-159.
Sherkat, D. E. and Ellison, C. G.: Structuring the Religion-
Environment Connection: Religious Influences on
Environmental Concern and Activism. Journal for the
Scientific Study of Religion, 46, 71-85 (2007).
Srinivasan, V. S. and Jafari, M. A. 1993. Fault
detection/monitoring using time petri nets. IEEE
Transactions on System, Man and Cybernetics, 23, 4,
1155-1162.
Wedel, M., Jensen, U., and Göhner, P. 2008. Mining
software code repositories and bug databases using
survival analysis models. In: 2nd ACM-IEEE
international symposium on Empirical software
engineering and measurement (Kaiserslautern,
Germany, October 09 - 10, 2008), pp. 282-284.
Yanaihara, N. et al. 2006. Unique microRNA molecular
profiles in lung cancer diagnosis and prognosis.
Cancer Cell, 9, 3, 189-198.
Yu, S. L. et al. 2008. MicroRNA Signature Predicts
Survival and Relapse in Lung Cancer. Cancer Cell,
13, 1, 48-57.
Zheng, Z., Lan, Z., Park, B. H., and Geist, A. 2009.
System log pre-processing to improve failure
prediction. In: 39th Annual IEEE/IFIP International
Conference on Dependable Systems & Networks
(Lisbon, Portugal, June 29 – July 2), pp. 572-577.
TOWARD A NON INVASIVE CONTROL OF APPLICATIONS - A Biomedical Approach to Failure Prediction
91
