
A HYBRID CLASSIFIER WITH GENETIC WEIGHTING
Benjam
´
ın Moreno-Montiel and Ren
´
e MacKinney-Romero
Departamento de Ingenier
´
ıa El
´
ectrica, Universidad Auton
´
oma Metropolitana, Iztapalapa, M
´
exico D.F., Mexico
Keywords:
Classification, Data Mining, Ensemble Based System, Knowledge Discovery in Databases (KDD), Machine
Learning.
Abstract:
This paper presents the results obtained when classifying a group of artificial and real world data, using a
Hybrid Classifier with Genetic Weighting (HCGW). The algorithm proposed is an ensemble based system, it
combines several types of classifiers: Naive Bayes, K-Means, k-Nearest Neighbours, C4.5, Decision Tables
and ADTree, using a voting criterion for weighted majority to combine the individual classifications of each
classifier, assigning the weights for each classifier using a genetic algorithm. We performed tests on data with
different tools for Data Mining, like SIPINA, TANAGRA and WEKA, to have a good comparison with the
proposed algorithm. Using standard measures such as accuracy, HCGW obtained better performance against
different implementations, from those tools, including traditional Ensemble Algorithms.
1 INTRODUCTION
One of the areas with a lot of interest in the last ten
years, is Data Mining, mainly due to the increase in
size of Data Bases (DB), with a resulting increase in
the potential of knowledge that lies within them. Data
Mining is an important phase in the process of Know-
ledge Discovery in Databases (KDD), performing ex-
ploration and analysis to identify nontrivial patterns
(knowledge) which are novel, potentially useful and
understandable in large DB. One of the main task
in Data Mining, is classification, used to predict the
class of an example within the data, and performed
by means of diverse types of classifiers.
When performing classification of data, we should
consider a model that allows us to classify each exam-
ple. Normally we have a data set used to build the
classification model, this is called the training set.
In which for each example its classification is given,
allowing us to obtain a trained model, thus enabling
us to classify new examples. These classification mo-
dels are called classifiers and can be found in the lit-
erature as decision trees, decision rules, classifiers
based in cases, neural networks, support vector ma-
chines, among many others.
In this work we propose to perform classification
of data using an ensemble based system. The objec-
tive of the ensemble based classifiers is to use several
types of classifiers to improve accuracy, using some
criterion to combine individual classifications. Bauer
et al.(Bauer and Kohavi, 1999), Schapire(Schapire,
2001), Quinlan(Quinlan et al., 2008) and others, con-
sider the construction of an ensemble, using weak
learners of only one kind of classifiers, usually deci-
sion trees, later a criterion to combine classifications
is applied, combining the individual classifications of
each weak learner, obtaining a model of classification
with better accuracy. Kelly et. al.(Kelly and Davis,
1991), consider the construction of an ensemble, us-
ing a classifier based on cases called k-nearest neigh-
bours, in which each one of the near neighbours is
weighted, using a genetic algorithm.
The algorithm we propose is called Hybrid Clas-
sifier with Genetic Weighting (HCGW). The HCGW
uses an ensemble based system of type Mixture of Ex-
perts and a weighted majority voting criterion to com-
bine the individual classifications of each classifier,
that is to say, each classifier has a different weight
according to the results of a genetic algorithm. This
algorithm provides a novel form to classification, be-
cause it actually considers several type of classifiers,
and not only decision trees, as it is normally found
in the literature. It also uses a new form to assign
weights to each classifier, unlike mixture of experts
neural network, using a genetic algorithm to assign
weights to each classifier. The main DB used for
this paper is a real world DB, used in the discovery
challenge for the European Conference on Machine
359
Moreno-Montiel B. and MacKinney-Romero R..
A HYBRID CLASSIFIER WITH GENETIC WEIGHTING.
DOI: 10.5220/0003460003590364
In Proceedings of the 6th International Conference on Software and Database Technologies (ICSOFT-2011), pages 359-364
ISBN: 978-989-8425-77-5
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

Learning and Principles and Practice of Knowledge
Discovery.
This paper is organised as follows. In Section 2
we will discuss previous work on ensemble based sys-
tems. In Section 3 we describe how we build the
HCGW. In Section 4 we will show the DB that was
used, so that in Section 5 the tests that were performed
to this DB and some standard data sets from the UC
Irvine repository(Frank and Asuncion, 2010), can be
discussed along with results obtained using several
tools of Data Mining. We will compare our results
with those from other tools used. We will perform a
statistical analysis to see the level of significance of
these tests using t-Test. Finally we will present some
Conclusions and Future Work.
2 PREVIOUS WORK
There are several methods for classification in Ma-
chine Learning, we will focus on ensemble based sys-
tems which we will review in this section:
• Bagging: Method introduced by
Breiman(Breiman, 1996), short for bootstrap
aggregating, is one of the earliest ensemble based
algorithms. This method is easy to implement,
the ensemble consists in taking a single type
of classifier (usually decision trees), generating
different models of the same classifier. A training
data subset is used to train a different classifier
of the same type, using 75% to 100% of the size
of DB. Finally individual classifications are then
combined by taking a majority vote.
• Boosting: In the 90’s this type of ensemble
based system was developed, by work made by
Schapire(Schapire, 2001), he proved that if a
weak learner is selected, and used with diffe-
rent sets, combining their individual classifica-
tions, it can be turned into a strong learner,
resulting in the Boosting Algorithm, considered
one of the seminal algorithms for Machine Lear-
ning(Polikar, 2006). The construction of the al-
gorithm is similar to the one of Bagging, a diffe-
rence being that it introduces the notion of sam-
ples with replacement for the phase of training
of weak learners. It also considers only decision
trees classifiers.
• Stacked Generalisation: This method was intro-
duced by Wolpert(Wolpert, 1992), using a set of
classifiers denoted by C
1
, C
2
, C
3
, ..., C
T
which
are trained first, so that an individual classification
for each of them is obtained, which are called the
First Level Base Classifiers. After obtaining these
individual classifications, a majority voting crite-
rion is selected, thus constructing the final classi-
fier, this phase is called Second Level Meta Clas-
sifier.
• Mixture of Experts: This method is similar to
Stacked Generalisation, it considers a set of clas-
sifiers denoted by C
1
, C
2
, C
3
, ..., C
T
, to per-
form first level base classifiers, later a classifier
C
T +1
combines the individual classifications of
each one considered, finding the final classifica-
tion. This model considers a phase in which the
weights are assigned to each classifier C
i
, i =
1,2,...,T , to finally apply a criterion of weighted
majority voting. Usually this part of the model is
performed by a neural network, called the gating
network(Polikar, 2006).
These are some approaches of how classification
can be performed in Data Mining using ensemble
based algorithms, they have been shown to be very
successful in improving the accuracy of classifiers for
artificial and real world DB, in this work we focused
on stacked generalisation using a weighted majority
voting criterion to combine class labels, in the next
Section our proposed algorithm is given in detail.
3 HYBRID CLASSIFIER WITH
GENETIC WEIGHTED (HCGW)
To construct any type of ensemble based systems,
three points are due to consider:
1. The fist point is to establish the number of clas-
sifiers that we will use, as well as the type of each
of them. This is seldomly done generally using
only one classifier such as decision trees.
2. The second point is the structure of the ensemble,
by means of which we will be able to group each
one of the classifiers, in the last section we saw
four different approaches for this.
3. Finally a criterion for combining the indivi-
dual classifications is chosen, majority voting or
weighted majority voting.
In this section we will describe how to construct
the HCGW, taking as reference the three points men-
tioned earlier. This is discussed in the following sub-
sections.
3.1 Number and Type of Classifiers
For this element of the HCGW we had to decide the
type, quantity and selection criterion of the different
ICSOFT 2011 - 6th International Conference on Software and Data Technologies
360

classifiers. In the literature we have a large number of
these, however we can not test every one of them, so
we took as an starting point the paper entitled ’Top 10
algorithms in data mining’ of J. Ross Quinlan (Quin-
lan et al., 2008), which presents the top ten algorithms
of classification.
Once we decided on which could be possible can-
didates, we had to use a selection criteria based on the
problematic we have, these criteria are listed below:
1. The implementation of a classifier should be sim-
ple.
2. Low running time.
3. Since it is an ensemble based system it is not
required to have a high percentage of accuracy,
since our objective is to gather various types of
classifiers to improve accuracy.
4. Finally, the classifiers selected, must support large
amounts of data.
After experimenting with classifiers from Quin-
lan’s paper and some others, we found that some clas-
sifiers do not meet the criteria that we set earlier, for
instance Support Vector Machine meet criteria one
and three, but two and four not fulfilled as it does not
support large amounts of data and the running time
is very high. We finally selected six classifiers that
meet the criteria. We have five supervised learning
(Naive Bayes, k-NN, Decision Tables, ADTree, C4.5)
and one unsupervised learning (K-Means) algorithms.
3.2 Structure of Ensemble
Having these six classifiers, we must establish the
structure for our ensemble based algorithm, we chose
mixture of experts, putting in one stack all the classi-
fiers. The selection of Mixture of Experts was due to
the fact that this type of ensemble based systems gives
us the chance to take the opportunity to use many dif-
ferent classifiers, which in Baggin and Boosting is not
used. This combined with a weighted voting approach
is a novel approach and the results showed that it is
good one.
3.3 Criterion of Combination of the
Individual Classifications
Each classifier considered, has different degrees of
accuracy, one of the characteristics of the models of
mixture of experts ensemble, and therefore we must
determine which criterion for combining the individ-
ual classifications to use, for constructing the classi-
fier C
T +1
.
As seen in the previous section for the mixture of
experts it is common to use neural networks, however,
neural networks have some issues, as the problem of
generalisation, in which the neural network learns the
training data correctly, but is not able to deal with to
new data. Another problem arises when using gra-
dient descent method to minimize the error, which
runs the risk of being trapped on local minimal and
not finding the best way to assign weights of classi-
fiers. To find the best form to weigh each classifier,
considering these problems, we use a different way
to assign them, which is genetic algorithm. To solve
the problem of being trapped in local minimum and
maximum, genetic algorithms have the genetic opera-
tor called mutation, which reduces the probability that
this occurs.
Since different weights give a different accuracy,
how can we know what is the best configuration? the
answer that we use was applying a simple genetic al-
gorithm, in which each population represents weights
for each of the classifiers. We chose six different
classifiers, thus the size of each chromosome in our
genetic algorithm was six. The codification of each
chromosome, has a specific weight in the range of [0,
0.5, 1, 1.5, 2, . . . , 4], defined arbitrarily.
In order to find the best combination of weights
assigned to each classifier, we must set the size of
the training and test set, for obtaining the individual
accuracy of each classifiers. Since we used a large
DB with a total of 379,485 records, a 10% random
sample is selected of the DB in order to avoid a long
runtime. This was selected since it was a good trade-
off between accuracy and runtime. It also falls in line
with statistical sampling. To do a simple random sam-
pling, as in our case, we have the following analysis
to obtain the sample size. Considering a confidence
level of 0.95, with a maximum error of 0.1 and a pi-
lot study gives a variance of 154.5, according to the
sample random simple calculation we have:
n
′
=
z
2
α/2
· σ
2
e
2
where:
• n
′
possible sample size,
• z
2
α/2
is the confidence level chosen,
• σ
2
population variance,
• e: maximum error,
If it is true that N > n
′
(n
′
−1), where N is the total
size of the data, it takes the value of n
′
as the sample
size, otherwise it will calculate a new sample size n,
as shown below:
A HYBRID CLASSIFIER WITH GENETIC WEIGHTING
361
n =
n
′
1 +
n
′
N
In this particular instance once we have the per-
formed the calculations we obtain a sample size of
51325. The value of the sample obtained represents
13.5% of the DB, we used 10% for practical reasons
and think is appropriate because it is near the value
obtained by statistical analysis.
Once the phase of training for each classifier is
finished, we obtain the individuals classifications, ha-
ving these we generate a population for the genetic
algorithm, where each chromosome represents a di-
fferent combination of weights. The genetic algo-
rithm is then executed a fixed number of iterations
using as the objective function to maximise accuracy,
using the weighted majority voting criterion to com-
bine class labels. The best combination of weights
found for this DB is in Table 1.
Table 1: Weights assigned to classifiers.
Case Name Weighted
1 Naive Bayes 3
2 ADTree 2.5
3 Decision Tables 1.5
4 C4.5 2
5 k-Nearest Neighbours 2
6 K-Means 2.5
3.4 Operation of HCGW
The operation of HCGW consists of the following
stages:
1. Training of the HCGW: First a random subset
of the DB is generated, to be able to begin with
the phase of training of each classifier considered.
Each of the classifiers is trained by a different
training set, selected randomly from the data base.
We use different training sets because they are tai-
lored for each classifier which is executed.
2. Configuration of Weights: A 10% test set is se-
lected (this percentage was used for the DB of
ECML PKDD, however this percentage can be
adjusted depending on the DB using). Once the
classification data is obtained, it is given to the ge-
netic algorithm which is executed for a fixed num-
ber of generations. This procedure is performed
only once.
3. Individual Classifications: The individual clas-
sifications for each classifier are obtained, consi-
dering the test set.
4. Combination of the Individual Classifications:
A weighted majority voting criterion is used to
combine class labels, so that for each of the exam-
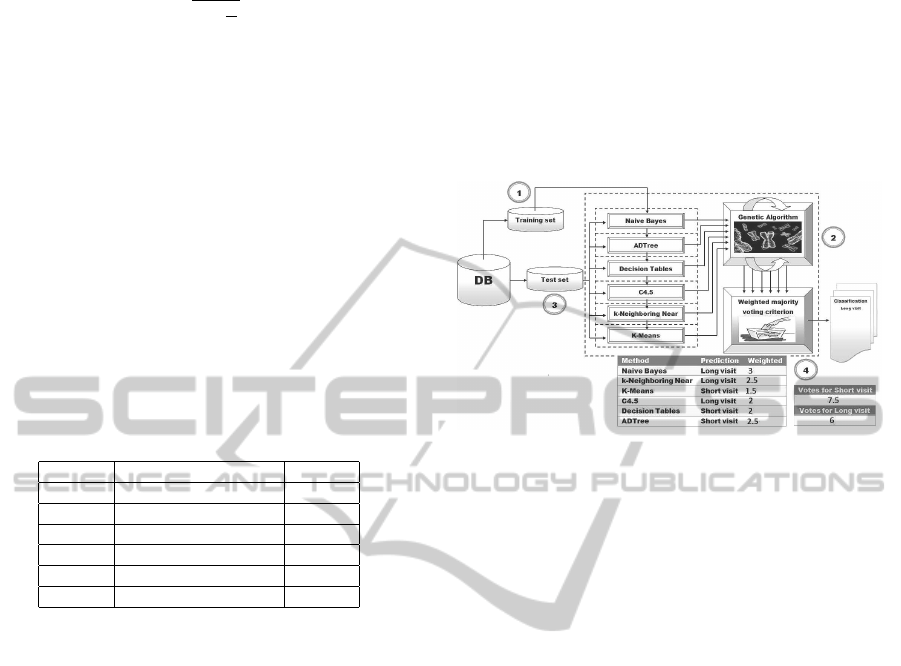
ples in the test set we get its classification. In Fi-
gure 1 we can observe the scheme of operation of
the HCGW with an example of how such classifi-
cation is performed.
Using our test DB we performed some experi-
ments with HCGW which are discussed in the Section
of Tests and Results.
Figure 1: Operation of the HCGW.
4 DB AND LEARNING TASK
As was mentioned before we use a DB which is from
ECML PKDD, this data was provided by the Gemius
Company, which is dedicated to the monitoring of In-
ternet on central and Eastern Europe. Within the DB
there were different problems to solve, but for this
work we only focus on one of them, which consists
in the following:
• The Length of the Visit. A visit is a sequence of
Page Views by one user. As web pages are identi-
fied by their categories, during one visit user may
view pages of one or more categories. Therefore
we define:
– Short visit: is a visit with page views of only
one category.
– Long visit: is a visit with pages views of two or
more categories.
The learning task is to answer the question
whether a given visit is short or long. The following
section will be dedicated to describe the experiments.
5 EXPERIMENTS
In this section we will review different experiments
performed on the DB. First, we will discuss the se-
lection of the test and training sets, the percentage
of the DB that we used to find the weights of the
ICSOFT 2011 - 6th International Conference on Software and Data Technologies
362
HCGW and the performance measures to consider.
Regarding the division of the data, in each experi-
ment we selected thirteen subgroups of random ele-
ments of the DB, where the distribution of the whole
DB was conserved, roughly 75% are short visits and
25% are long, having a size of 5,000, 10,000, 20,000,
40,000, 80,000, and so on in multiples of 40,000 until
360,000, the last set is the full DB (379,485 records)
These thirteen subgroups conformed our test and
training set, that is to say, for the training set, the class
of each one of the examples is conserved, which is the
type of visit. For the test set, we eliminated the class
of each example, conserving the other attributes.
In the experiments carried out with the DB of
EMCL PKDD, there are only on two classes due to
the structure of the DB, but since implementations of
each component are our own, data can be classified
with more than two classes.
Once we had these sets, we selected the data for
the genetic algorithm, in order to find the weights of
each one of the classifiers. We must take into account
the great number of iterations due to the size of the
population, for example, if we have 10,000 examples
that were classified of individual way by the six clas-
sifiers, we will have a matrix 6 X 10,000. For this
matrix we must test each chromosome, to see what
which is the final classification with that combination,
and thus repeat until obtaining the function of aptitude
of each chromosome.
Within machine learning there are different per-
formance measures, in this paper we only show the
results of Accuracy. Accuracy is the percentage of
examples classified correctly in the test set.
In the following section we will present the re-
sults found when performing these experiments with
the thirteen sets, comparing different tools against the
HCGW, showing the accuracy results obtained.
6 RESULTS
The following tools for Data Mining(Witten and
Frank, 2005): SIPINA(Witten and Frank, 2005),
TANAGRA(Witten and Frank, 2005), WEKA (Wit-
ten and Frank, 2005) and our own implementations in
Matlab R2008b (7.7), were employed, and they were
used in the classification of the Gemius DB.
We selected different sizes of training set and the
thirteen subgroups, to be able to perform classifica-
tion with the four previous tools. Were tested with
a set of classifiers for each tool consideration, how-
ever for this paper those which showed the best results
are Ensemble Based Systems so we focused on them.
First we will show the results for ensemble based sys-
tems of tool WEKA and then we will select the best
tools to compare against HCGW.
For ensemble based systems we selected the tool
WEKA, since it has a large number of algorithms
implementing this type of classifier, and its imple-
mentations gave the best accuracy, the results with
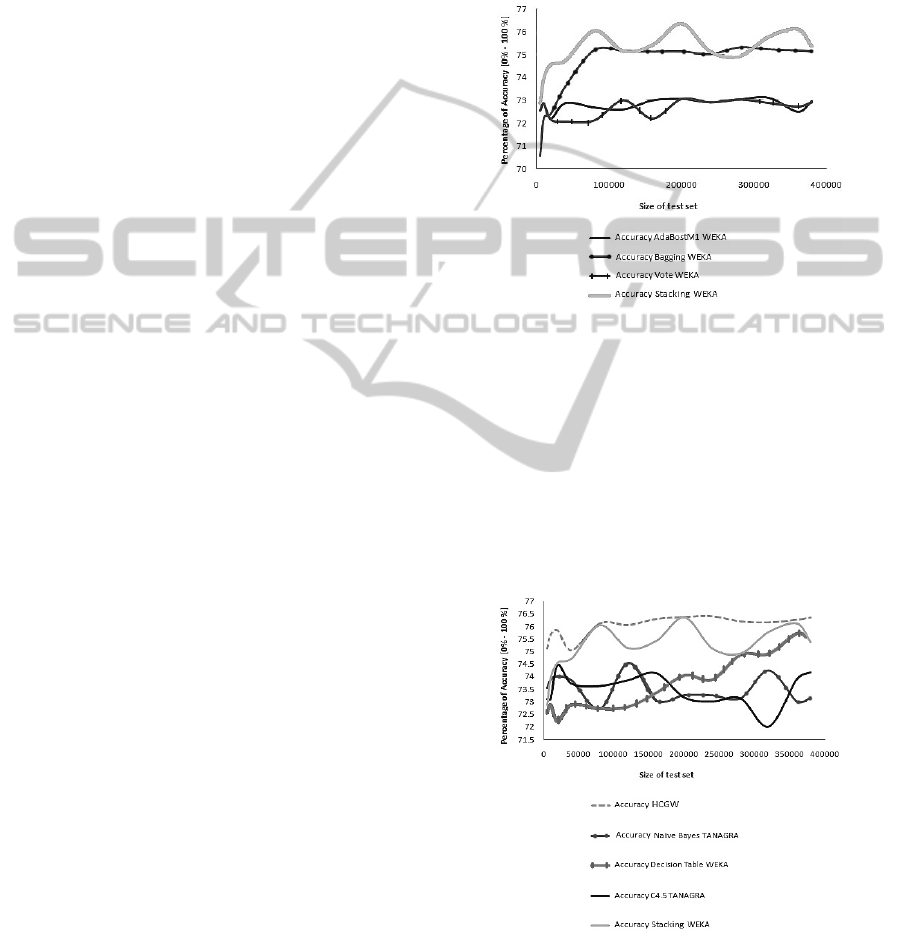
Ensemble Methods are shown in Figure 2.
Figure 2: Comparison of accuracy of Ensemble Methods.
In Figure 2 the Stacking method shows the most ac-
curacy, this method is similar to our HCGW, it is a en-
semble methods of type Stacked Generalisation (vot-
ing by majority), but it only considers decision trees
to classify, and not several different methods as we
do. The tool achieves a 75.19% in accuracy. Finally
we calculated the accuracy for our HCGW, and com-
pared the results for each one of the thirteen sets, with
the best results previously obtained, Figure 3 shows
these results.
Figure 3: Comparison of accuracy with HCGW.
As we can see in Figure 3 the accuracy grew
2.26% with respect to the other methods considered
for the tests, this gives us the result that the HCGW
performs better than traditional techniques.
A HYBRID CLASSIFIER WITH GENETIC WEIGHTING
363

Once these results with this DB were obtained, we
did a t-Test, taking accuracy from the HCGW and
the one from Stacking of WEKA, obtaining a level
of significance high since we are confident with a
99.9995% that the results of our model are signifi-
cantly different and better than those than we obtained
with Stacking of WEKA.
Figure 4: Comparison of times.
In Figure 4 we can see the runtimes of the tools
we used, we can observe that Stacking of WEKA and
HCGW are those that take more time. This is ex-
pected as they use several classifiers, the complexity
of our algorithm is approximately equal to the sum
of the individual complexities of the classifiers used.
The results obtained with this DB and some results
with the UC Irvine repository(Frank and Asuncion,
2010), are shown in Table 2:
Table 2: Final comparison of results.
Name Records HCGW Stacking (WEKA)
Gemius complete 379485 76.03 75.19
Credit (German) 1000 72.33 71.75
Mushroom 8124 92.46 95.63
Australian 690 84.12 82.75
In the Table 2 we can observe that for DB
Gemius complete, Credit and Australian, the accu-
racy of the HCGW is better than the methods of tool
WEKA, which were chosen because they have bet-
ter accuracy than other tools. For Mushroom, Stack-
ing obtains a better accuracy which can be explained
since is best fitted to a decision tree method as done
by WEKA classifier has better results than us and this
was adjusted in a better way to this DB has a small
size, the No free lunch(Wolpert and Macready, 1997)
theorem applies here, since there is no classifier that
is the best for all the problems.
7 CONCLUSIONS AND FUTURE
WORK
This paper presents an ensemble based algorithm of
type stacked generalisation, taking several types of
classifiers and implements a weighted majority vot-
ing criterion to combine class labels, using a genetic
algorithm to assign the weights each classifier. This
model of classification we called it a Hybrid Classi-
fier with Genetic Weighting, which is a novel algo-
rithm, because it actually considers several type of
classifiers, and not only decision trees like normally
found in the literature. It uses as well a genetic algo-
rithm for the allocation of weights. With this model of
classification, we obtained a better accuracy for each
one of the tests we made to the DB gemius complete,
comparing it with different methods from other tools.
Since running time is a major issue as future work we
will look into parallel computing as means to solve it.
There would be parallel versions of each classifier, the
genetic algorithm as well as the HCGW component
that handles the combination of individual classifica-
tions. This, we believe, would lower the total running
time allowing larger data sets to be handled as well as
being able to consider some other classification algo-
rithms which were too costly for this work.
REFERENCES
Bauer, E. and Kohavi, R. (1999). An Empirical Comparison
of Voting Classification Algorithms: Bagging, Boost-
ing, and Variants., volume 36. Kluwer Academic Pub-
lishers.
Breiman, L. (1996). Bagging Predictors., vol. 25. Kluwer
Academic Publisher.
Frank, A. and Asuncion, A. (2010). UCI machine learning
repository.
Kelly, J. D., J. and Davis, L. (1991). A hybrid genetic
algorithm for classification. In Proceedings of the
Twelfth International Joint Confirence on Artificial In-
telligence., pages 645–650.
Polikar, R. (2006). Ensemble based systems in decision ma-
king. IEEE Circuits and Systems Magazine 6:21-45
Quinlan, J. et al. (2008). Top 10 algorithms in data mining.
Knowledge and Information Systems., 14:1–37.
Schapire, R. (2001). The boosting approach to machine
learning: An overview. AT&T Labs Research Shan-
non Laboratory.
Witten, I. and Frank, E. (2005). Data Mining: Practi-
cal Machine Learning Tools and Techniques. Morgan
Kaufmann Publishers. 2nd edition.
Wolpert, D. (1992). Stacked generalization. Neural Net-
works., 5:241–259.
Wolpert, D. and Macready, W. (1997). No free lunch the-
orems for optimization. Evolutionary Computation,
IEEE Transactions on, 1:67–82.
ICSOFT 2011 - 6th International Conference on Software and Data Technologies
364
