
INFRASTRUCTURE FOR METAGENOME DATA
MANAGEMENT AND ANALYSIS
Tatiana Tatusova
National Center for Biotechnology Information, National Library of Medicine
National Institutes of Health, 9600 Rockville Pike, Bethesda, MD, 20892, U.S.A.
Keywords: Database, Sequence analysis, Metagenomics.
Abstract: Metagenome sequencing projects are generating unprecedented amounts of data. Public sequence archive
databases are challenged with large-scale data management issues including data storage, quick search and
retrieval of the sequence data for further analysis. The sequence data is linked to the rich set of metadata
attributes such as geochemical and ecological parameters for environmental projects and clinical patient in-
formation for human microbiome studies. That complex collection of heterogeneous information has to be
integrated, organized and presented to the users in a meaningful and the most useful way. For the last 20
years The National Center for Biotechnology Information (NCBI) has been developing the infrastructure
that allows an easy storage and distribution of various types of bimolecular data as well as data integration
and easy navigation in complex information space. Here we describe NCBI resources that are used for me-
tagenomics data management.
1 INTRODUCTION
New generation sequencing technology made it
possible to study microbial communities in their
natural environment. By collecting samples directly
from the environment and sequencing DNA without
isolation and growing in the artificial conditions
researches are given an opportunity to understand
the role of microbial organisms in ecological sys-
tems. The questions the researches are usually ask
are:
1) what is the structure of microbial community and
relative abundance of different species?;
2) What is the functional role of the bacterial com-
munities in the ecosystem? In other words scientists
want to know “who they are?” and “what they do?”
One well established way to answer the first ques-
tion is to collect and sequence 16S RNA genes and
perform phylogenetic analysis. To answer the
second question genomic DNA, assembled and an-
notated. The analysis of the predicted proteins might
provide some insight into the functional role of bac-
terial communities in the regulation of biochemical
processes in ecosystems. More recently with the
RNA Seq technology metatranscriptome data be-
came available for the analysis the expression level
of functional activity of microbial communities.
Sequence data generated by metagenome
projects is made available to the research community
through public data achieves described in section 1.
Sequence data by itself doesn’t contain enough
information for the analysis, it is necessary to frame
the physic-chemical context within which the data is
to be interpreted. The initial step in any metagenom-
ics study requires the collection of samples destined
for analysis. The geo-chemical or medical characte-
ristics describing a sample constitute the "meta data"
intricately tied to a given sample and aid in interpret-
ing the biological significance of the genetic infor-
mation. Linking the metadata to the sequence data
extracted from the sample is one of the key elements
in further analysis. Computational analysis of the
metagenomics creates a great challenge due to the
huge volume of the metagenomics data requiring
extremely powerful computational resources and
novel approaches to sequence analysis and visualiza-
tion methods.
NCBI has recently developed new resources that
allow capturing some metadata associated with se-
quence submission such as the description of the
project, description of each sample and geochemical
and ecological data associated with the study. Sec-
tion 2 will provide the detailed description of the
specialized resources.
357
Tatusova T..
INFRASTRUCTURE FOR METAGENOME DATA MANAGEMENT AND ANALYSIS.
DOI: 10.5220/0003333803570362
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (Meta-2011), pages 357-362
ISBN: 978-989-8425-36-2
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

In addition to general archive databases NCBI
has created specialized resources and tools that can
be utilized in metagenomics data analysis. These
resources are discussed in section 3.
2 PRIMARY DATA ARCHIVES
As a national resource for molecular biology infor-
mation, National Center for Biotechnology Informa-
tion develops, distributes, supports, and coordinates
access to a variety of databases and software for the
scientific and medical communities (Sayers, 2010).
2.1 Sequence Read Archive
The advent of massively parallel sequencing tech-
nologies has opened an extensive new vista of re-
search possibilities — elucidation of the human
microbiome, discovery of polymorphisms and muta-
tions in individual genomes, mapping of protein–
DNA interactions, and positioning of nucleosomes
— to name just a few. In order to achieve these
research goals, researchers must be able to effective-
ly store, access, and manipulate the enormous vo-
lume of read data generated from massively parallel
sequencing experiments.
In response to the research community’s call for
such a resource, NCBI, EBI, and DDBJ, under the
auspices of the International Nucleotide Sequence
Database Collaboration (INSDC), have developed
the Sequence Read Archive (SRA) data storage and
retrieval system (Shumway, 2010). The SRA not
only provides a place where researchers can archive
their sequence read data, but also enables them to
quickly access known data and their associated ex-
perimental descriptions (metadata).
Now that the archive has reached an initial state
of completion and is publically available at NCBI, it
is being deployed at EBI (under the name European
Read Archive, or ERA), and soon will also be dep-
loyed at DDBJ (under the name DDBJ Read Arc-
hive, or DRA). NCBI and EBI have already begun
exchanging data, and once the DRA is in place at
DDBJ, there will be a regular data exchange be-
tween all three INSDC members.
In order to store and retrieve the enormous
amount of data generated by massively parallel se-
quencing technologies, NCBI, EBI and DDBJ
needed to create a data repository that has much of
the power of a relational database while being
lightweight, transportable and flexible like flat-file
storage. The solution was to create a hybrid relation-
al database with a file-based and column-oriented
design.
Within SRA the data are organized into four
types of records: studies (SRP accessions), experi-
ments (SRX accessions), samples (SRS accessions)
and runs (SRR accessions). Studies contain one or
more experiments, each of which contains one or
more runs, each of which in turn may contain data
on tens of millions of individual reads. The various
record types representing data from a study are all
linked to one another within Entrez
(www.ncbi.nlm.nih.gov/sra/), allowing users to
browse the data easily on the web.
2.2 GenBank – Nucleotide Sequence
Archive Database
GenBank (Benson et al., 2010) is a comprehensive
database that contains publicly available nucleotide
sequences for more than 300 000 organisms named
at the genus level or lower, obtained primarily
through submissions from individual laboratories
and batch submissions from large-scale sequencing
projects, including whole genome shotgun (WGS)
and environmental sampling projects. NCBI builds
GenBank primarily from the submission of sequence
data from authors and from the bulk submission of
expressed sequence tag (EST), genome survey se-
quence (GSS) and other high-throughput data from
sequencing centers. GenBank data is available at no
cost over the Internet, through FTP and a wide range
of web-based retrieval and analysis services.
2.3 Metadata: BioProject and
BioSample
BioProject. New technologies have significantly
increased the volume of data that can be generated
and submitted to archival database resources. Ge-
nome project is no longer limited to the genome
sequencing, assembly, and annotation. New types of
experimental studies include epigenomics, proteo-
mics, metabolomics and more ’omics’. Advances in
sequencing technologies have also changed the
scope of genomic studies; it became possible to
sequence multiple genomes of many different organ-
isms starting from hundreds of bacterial strains to
1000 human individuals. It is also possible to se-
quence microbial populations in their natural envi-
ronment without growing them in culture but by
sequencing the samples collected from the environ-
ment. Our view on genomic, metagenomics and
biomedical projects is rapidly changing. That affects
the way the data is organized and represented in
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
358

NCBI databases. BioProject database provides a
mechanism to access datasets that are otherwise
difficult to find. The definition of a set of related
data, a ‘project’ is flexible and supports the need to
define a complex project and various distinct sub-
projects using different parameters.
BioSample. The time, place and collection method
can profoundly affect the microbial composition in a
sample. Geographical location, biochemical charac-
teristic of the natural habitat, ecological and clinical
information needs to be captured and linked to the
sample data during the submission of the raw data. It
is highly important to develop a set of uniform stan-
dards for sample information to make future com-
parisons between different data sets easier and so
provide greater biological insight.
NCBI new BioSample database (http://www.
ncbi.nlm.nih.gov/biosample) is meant to support
sample descriptions and standard attributes for all
biological samples.
The new database provide a good infrastructure
for future submissions of sample information but a
common set of standard attributes is yet to be devel-
oped.
Example of BioSample record in Entrez:
Soil metagenome SRA sample SRS009922
Identifiers
SRA:SRS009922
Organism
Soil metagenome
unclassified sequences; metagenomes; ecological metagenomes
Attributes
No attributes
Submitter
JGI
Description
The tropical forest soil sample used for metagenome se-
quencing was collected in a subtropical lower montane
wet forest in the Luquillo Experimental Forest (18.30N,
65.83W), which is part of the NSF-sponsored Long-Term
Ecological Research program in Puerto Rico. The climate
in this region is relatively aseasonal, with mean annual
rainfall of 4500 mm and mean annual temperatures of
22C to 24C. Soils were collected from the Bisley wa-
tershed approximately 250 meters above sea level from
the 0-10 cm depth using a 2.5 cm diameter soil corer.
Sampling date: Summer 2008
ID: 8167
The new database provide a good infrastructure
for future submissions of sample information but a
common set of standard attributes is yet to be devel-
oped.
2.4 Developing Community Standards
for Metagenome Data
The astonishing increase in the amount of data gen-
erated by metagenomics projects that involve shot-
gun sequencing of all the organisms in an environ-
mental sample creates an unanticipated situation in
the field. Data storage and retrieval is becoming a
problem for current database designs, and compre-
hensive analysis of the metagenomics data, which is
far more complex analysis of a genome, is becoming
computationally intractable with existing resources
and pipelines. (see Nature Methods 6, 623 (2009)).
A single lab can no longer alone perform a compre-
hensive analysis of metagenomics data. The devel-
opment common standards would facilitate the data
exchange, sharing and comparisons of the results
across different groups.
The recently formed M5 (metagenomics, meta-
data, meta analysis, multi-scale models and meta
infrastructure) Consortium will be proposing a
promising solution, the 'M5 Platform', later this year.
The success of developing standards depends on the
ability of the public repositories and biologists gene-
rating the data to agree on common data models and
unified data formats. There is commitment, howev-
er, from GenBank, the European Molecular Biology
Laboratory's Nucleotide Sequence Database, and the
DNA Databank of Japan, to capture the metadata
and associate it with the genome records, in the
sequence records and in a project description.
3 REFERENCE SEQUENCE
COLLECTION
NCBI's Reference Sequence (RefSeq) is a public
database of nucleotide and protein sequences with
feature and bibliographic annotation. For more de-
tails see (Pruit et al, 2009).
3.1 Reference Microbial Genomes
Reference collection of complete microbial genomes
includes complete annotated genomes that can be
used as standards for microbial genome annotation,
WGS (Whole Genome Shotgun) genomes that
represent major taxonomic group in the absence of a
complete genome.
3.2 Reference Targeted Loci
Reference collection of targeted loci includes tar-
geted sequence regions that support specific report-
INFRASTRUCTURE FOR METAGENOME DATA MANAGEMENT AND ANALYSIS
359

ing or identification needs; for example, gene-
specific benchmarks that are used for identification
purposes. The small subunit ribosomal RNA (16S in
prokaryotes and 18S in eukaryotes) is a useful phy-
logenetic marker that has been used extensively for
evolutionary analyses. This project is the result of an
international collaboration with a number of ribo-
somal RNA databases that curate and maintain se-
quence datasets for these markers. The initial scope
of the project is to compare curated 16S markers that
correspond to type strains and near full length se-
quences from all contributing databases. Sequences
and taxonomic assignments that are in agreement in
all databases will have Reference Sequence records
corresponding to the original GenBank record. The
RefSeqs may contain corrections to the sequence or
taxonomy as compared to the original INSD submis-
sion, and may have additional information added
that is not found in the original. The Refseq Tar-
geted Loci web resource http://www.ncbi.
nlm.nih.gov/genomes/static/refseqtarget.html con-
tains comparison tool for different outside resources
of targeted loci data. One of the goals of the project
is to create a unique reference set that can be used by
many existing databases. The data are available for
download at NCBI ftp site ftp://ftp.ncbi.nih.
gov/genomes/TARGET/
4 ANALYSIS TOOLS
AND RESOURCES
4.1 Family of Standard BLAST
Programs
The BLAST programs (Altschul et al., 1990;
Altschul et al., 1997; Ye et al., 2006) perform se-
quence-similarity searches against a variety of nuc-
leotide and protein databases.
A special search program for genomic and meta-
genomic data MegaBLAST (Ye et al., 2006), is a
faster version of standard nucleotide BLAST de-
signed to find alignments between nearly identical
sequences, typically from the same species. It is
available through a separate web interface that han-
dles batch nucleotide queries and can be used to
search the rapidly growing Sequence Read Archive
as well as the standard BLAST databases. For rapid
cross-species nucleotide queries, NCBI offers Dis-
contiguous MegaBLAST, which uses a nonconti-
guous word match (Zhang et al., 2000) as the nuc-
leus for its alignments. Discontiguous MegaBLAST
is far more rapid than a translated search such as
blastx, yet maintains a competitive degree of sensi-
tivity when comparing coding regions. Sequence
read BLAST searches are now offered for transcript
and whole genome sequence data sets from 454
Sequencing systems, and regular expression pattern
matching against short reads of all types is now
possible.
4.2 Customized BLAST Databases
4.2.1 SRA BLAST
SRA data are rapidly dominating all other sequence
data. Already the number of DNA bases available in
SRA exceeds the number of bases in GenBank. In
fact the output of a single important project, the
1000 genomes project (www.1000genomes.org),
will produce more than 25 times the number of bases
that are currently in GenBank by the time the project
is completed. The NCBI and SRA will continue to
support submission, retrieval, and analyses of these
increasingly challenging and complex sequencing
data. Means of displaying data, analyses, and inte-
gration of SRA data with other molecular databases
will continue to improve making the SRA data a
prominent part of the discovery system at the NCBI.
In addition to text searches of the SRA experi-
ments through Entrez, NCBI also offers a nucleotide
BLAST service for sequence similarity searching of
454 sequencing reads for transcriptome studies. This
service is accessible from the “Specialized BLAST”
section of the BLAST Homepage.
4.2.2 Genomic BLAST
Genomic BLAST (Cummings et al., 2002), a novel
graphical tool for simplifying BLAST searches
against complete and draft genome assemblies. This
tool allows the user to compare the query sequence
against a virtual database of DNA and/or protein
sequences from a selected group of organisms with
finished or unfinished genomes. The organisms for
such a database can be selected using either a graph-
ic taxonomy-based tree or an alphabetical list of
organism-specific sequences. The first option is
designed to help explore the evolutionary relation-
ships among organisms within a certain taxonomy
group when performing BLAST searches.
4.2.3 Concise BLAST
The vast increase in genomic sequences has led to a
flood of data to the protein databases as well. Many
strain-specific genomes are now being sequenced
(for example Streptococcal genomes). The result can
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
360
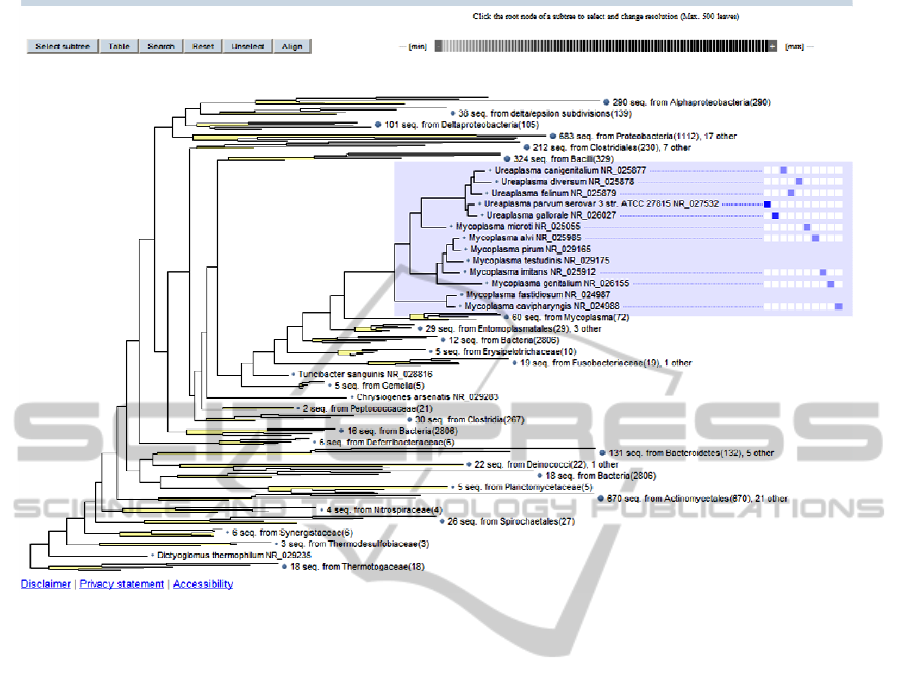
Figure 1: 16S Ribosomal RNA Reference Sequence Similarity Search Beta release. This tool visualizes BLAST results of
the query sequence search by mapping them on a phylogenetic tree. Query: >NC_002162 |:145338-146803|16S ribosomal
RNA| [gene=rRNA_16S-1] [locus_tag=UUr01].
be an overwhelming amount of data to look through
when executing BLAST similarity searches. In order
to help alleviate both the processing of the data and
to present a broader taxonomic view, the concise
protein database was constructed. Web interface can
be accessed from Microbial Genomes home page or
directly at http://www.ncbi.nlm.nih.gov/genomes/
prokhits.cgi.
The database is constructed from the clusters of
related proteins [ref] Clusters may span a large tax-
onomic branch (kingdom) or may reside at a specific
node (family, genus, species, etc.). Clusters may
consist of many proteins, or be comprised of only
two proteins. From this entire set of clusters, genus-
specific clusters are used for this database. From the
proteins at the genus-level, one (randomly selected)
is chosen as a representative for the Concise Micro-
bial Protein BLAST database and will be found in
BLAST queries. The other proteins in the cluster are
automatically linked to this representative and will
also be found in the search results, although without
the BLAST score and E-value as they are not specif-
ically examined. All proteins that do not belong to
the genus-level clusters are also added to the data-
base for completeness. The result is faster
processing times and reduced load on the database.
The broader taxonomic view will help eliminate
some of the redundancy that is found when many
proteins of closely related organisms are found in
BLAST results.
4.3 Analysis of 16S Ribosomal
RNA Data
Similarity search give you a list of the 10 top
BLAST hits as well as the position of the hits on the
phylogenetic tree. Coursing tree visualization algo-
rithms developed at NCBI allow showing trees for
large datasets with various levels of the resolution.
5 CONCLUSIONS
NCBI provides a basic infrastructure for the se-
quence data and a framework for metadata that de-
scribes project, study, and sample. However, com-
mon standards for the metadata and a new data mod-
el for metagenome sequence data have yet to be
INFRASTRUCTURE FOR METAGENOME DATA MANAGEMENT AND ANALYSIS
361

developed. A special interest group (SIG M3) at
ISMB meeting had brought together researchers
collecting samples for metagenomic analysis with
those building the computational infrastructure re-
quired to fully exploit them with those thinking
about the implementation of standards. This discus-
sion initiated by Genomic Standards Consortium -
GSC
(http://gensc.org/gc_wiki/index.php/Main_Page) is a
good start towards developing standards for metage-
nome data that will be supported by major databases
and utilized through already existing NCBI infra-
structure.
REFERENCES
Sayers E. W. et al.: Database resources of the National
Center for Biotechnology Information. Nucleic Acids
Res. 2010 Jan; 38 (Database issue): D5-16.
Shumway M.: The Sequence Read Archive (SRA) – A
worldwide resource. Nucleic Acids Res. 2010 Jan; 38
(Database issue): D.
Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J.,
Sayers E. W.: GenBank. Nucleic Acids Res. 2010 Jan;
38 (Database issue): D46-51.
Pruitt K. D., Tatusova T., Klimke W., Maglott D. R.:
NCBI Reference Sequences: current status, policy and
new initiatives. Nucleic Acids Res. 2009 Jan; 37 (Da-
tabase issue): D32-6.
Altschul S. F., Gish W., Miller W., Myers E. W., Lipman
D. J.: Basic local alignment search tool. J. Mol. Biol.
1990; 215: 403-410.
Altschul S. F., Madden T. L., Schaffer A. A., Zhang J.,
Zhang Z., Miller W., Lipman D. J.: Gapped BLAST
and PSI-BLAST: A new generation of protein data-
base search programs. Nucleic Acids Res. 1997;
25:3389-3402.
Ye J., McGinnis S., Madden T. L.: BLAST: improvements
for better sequence analysis. Nucleic Acids Res. 2006;
34: W6-W9.
Zhang Z., Schwartz S., Wagner L., Miller W.: A greedy
algorithm for aligning DNA sequences. J. Comput.
Biol. 2000; 7: 203-214.
Cummings L., Riley L., Black L., Souvorov A., Resen-
chuk S., Dondoshansky I., Tatusova T.: Genomic
BLAST: custom-defined virtual databases for com-
plete and unfinished genomes. FEMS Microbiol Lett.
2002 Nov 5; 216 (2): 133-8.
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
362
