
Content-Based Computer Tomography Image Retrieval
on a Whole-Body Anatomical Reference Set:
Methods and Preliminary Results
Aur
´
eline Quatrehomme
1,2
, Denis Hoa
1
, G
´
erard Subsol
2
and William Puech
2
1
IMAIOS, Cap Omega - CS 39521, Rond Point Benjamin Franklin
34960 Montpellier Cedex 2, France
2
Universit
´
e Montpellier 2 / CNRS, LIRMM, 161 rue Ada, 34095 Montpellier Cedex 5, France
Abstract. This paper describes a CBIR system presenting two key points: a
generic CT data, as well as a novel algorithm for combining visual features. The
descriptors express grey levels, texture and shape of the images. A normalization
method is proposed in order to improve the quality of indexing and retrieval. Our
selected features and our combination method are effective for retrieving images
from a whole-body reference set.
1 Introduction
The number of numerical images produced increases every day, particulary in medical
imaging, which has benefited from recent technologic improvements. Strong needs for
storing, indexing and retrieving these huge amounts of data have emerged at the same
time.
A Content-Based Image Retrieval (CBIR) system aims to retrieve the most similar
images to a query from a database. In CBIR systems, the query is an image, as textual
queries cannot describe precisely all the visual characteristics of an image. The main
idea is to extract some ”features” from the images, which will be compared for retrieval.
Figure 1 presents the general framework of CBIR systems, which processes in two
phases. The first one extracts some visual descriptors from the images in the database
and stores them. The second one is the real-time retrieval phase. The user inputs a query
image, from which descriptors are extracted and compared to the ones in the features
database. The system finally retrieves most similar images.
In the field of medical practice, CBIR is often associated to Computer Aided Diag-
nosis (CAD). By integrating computer assistance in the diagnosis process, the goal is
not to get rid of medical expertise but to improve its efficiency and accuracy. Current
trend is to design diagnosis-driven (and then very specific) systems, which makes their
evaluation a significant problem.
Our aim is to create a Content-Based Radiology Image Retrieval (CBRIR) system
which retrieves a close positioning of a medical image in the body (in order to know
if the image content is the brain, the liver...). Image positioning is the first step of ra-
diology diagnosis. Most systems work on very specific data, as mammographies [1] or
inter-vertebrae disks [2]. This was defined in [3] as the ”use context gap”. The system
we propose covers the full human body. The remainder of this paper is divided into
Quatrehomme A., Hoa D., Subsol G. and Puech W..
Content-Based Computer Tomography Image Retrieval on a Whole-Body Anatomical Reference Set: Methods and Preliminary Results.
DOI: 10.5220/0003312500520061
In Proceedings of the 2nd International Workshop on Medical Image Analysis and Description for Diagnosis Systems (MIAD-2011), pages 52-61
ISBN: 978-989-8425-38-6
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)
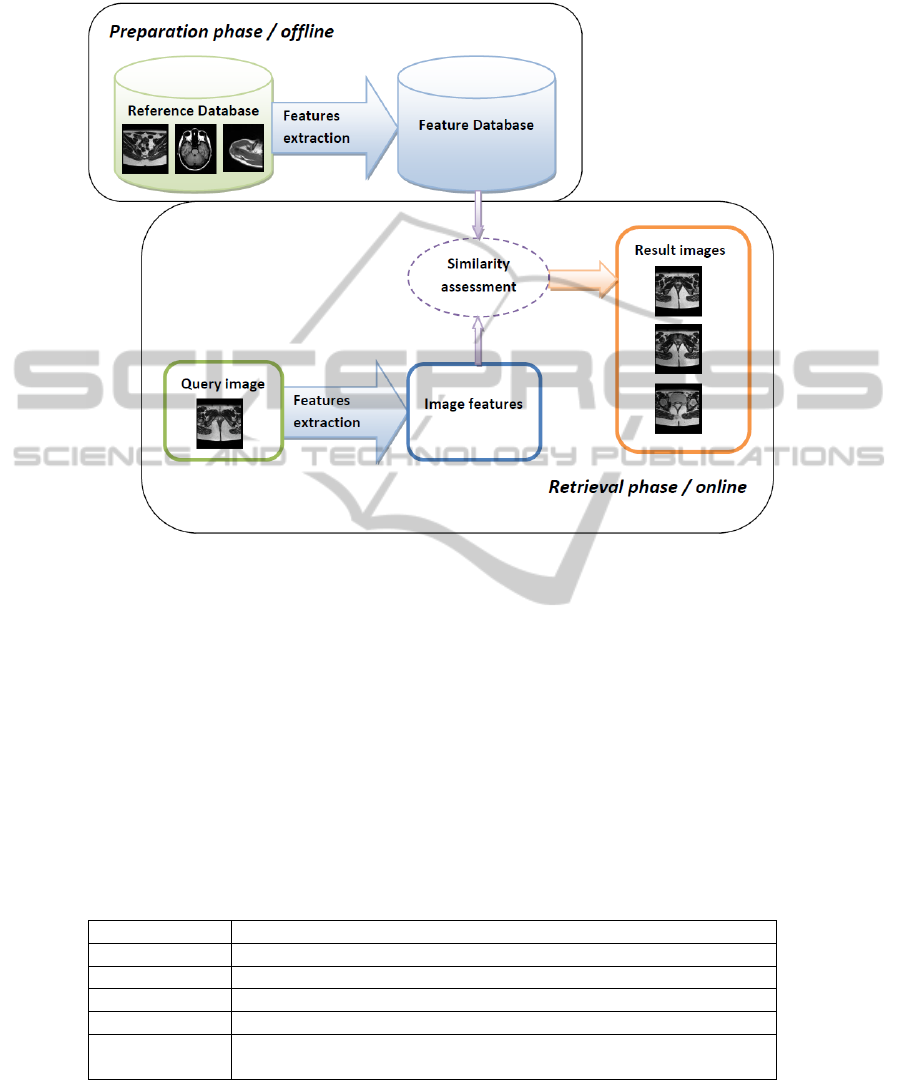
Fig. 1. General framework CBIR systems.
tree sections. In Section 2, we describe the characteristics of our system, our data and
method, including our features combination algorithm. In Section 3, we present results,
and in Section 4, we state the conclusion of this work.
2 Material and Methods
2.1 Our CBIR System Characteristics
In a previous paper [4], we presented an overview of the key points of CBIR systems.
Table 1 shows the characteristics of the system we propose.
Table 1. Characteristics of proposed CBIR system.
Image Modality Computer Tomography (CT)
Data content General (from head to pelvis)
Application CT slice positionning in the body
Query A single image
Visual features Descriptors used for expressing the image content (described Section 2.4).
Distance measure
In order to express the similarity / dissimilarity between two images. (de-
scribed afterwards)
53
2.2 Our Image Database
We work on an anatomical atlas, proposed by the company IMAIOS (e-Anatomy). The
anatomical structures are localized and captionned in each image of a huge set of over
20,000 CT images. Our reference set is made of 380 CT images with a 3 mm section
thickness, from a single patient. It goes from the brain to the pelvis. Thus, even if the
images are in 2D dimension, 3D information can be extracted. The originality of our
approach is due to this volumic information and the cover of a large range instead of
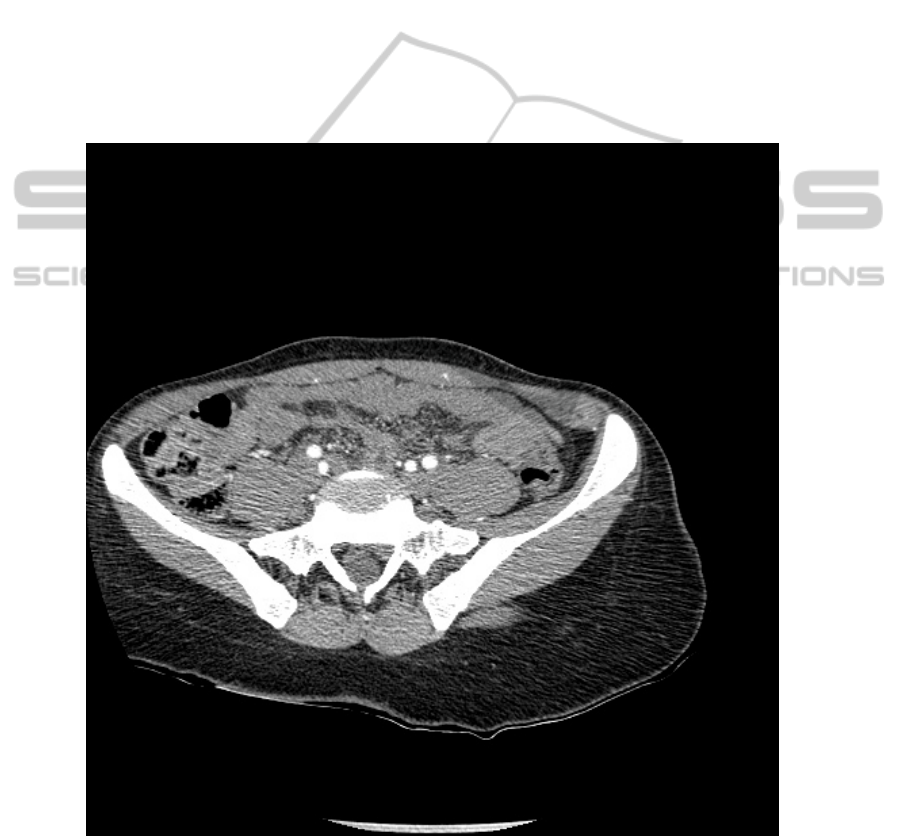
focusing on a single anatomical structure. Our test database is a set of 10 CT images
coming from different patients than the one used as reference. The aim of our system
being to work in a real medical context, the test images were not chosen ideal: some
elements as table or pipes, external to the patient, can be seen on them, the body is not
always centered. A test image is shown on Fig. 2.
Fig. 2. One of our test images: patient badly positioned, and visible examination table.
54
2.3 Normalization
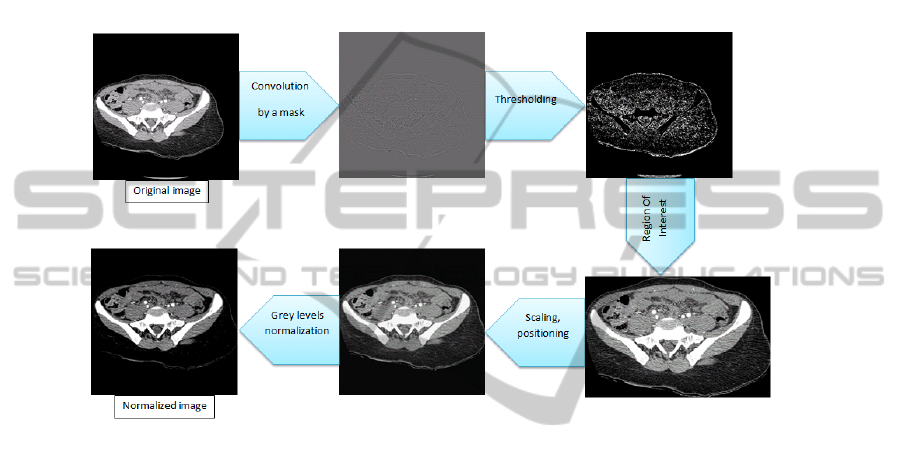
In order to work on the significant part of the CT images, we implemented a normaliza-
tion method. First of all, the original image is convolued by a mask in order to get the
contours. Then a threshold is applied, before getting rid of unwanted elements on the
image (for example external to the patient body: table, pipes...). The Region Of Interest
(ROI) containing the patient is automatically defined, then is being scaled and posi-
tioned in a 256x256 image. The grey levels are finally normalized. Figure 3 presents
the normalization process on an image of our test dataset.
Fig. 3. Normalization process.
2.4 Features and Distance Measures
Visual features, also named descriptors, express the image content. A presentation of
the different types of features used in recent CBRIR systems and their classification
were presented in our previous paper [4], reviews as [5, 6] may be read for a more
global view of existing CBIR systems. We will list here the one we currently tested.
In this work, we use only general descriptors, that can be extracted from any image.
The features describe either color, texture or shape. We compute them over the whole
image, or on each block obtained by dividing the image in small patches of equal size.
Color Features: Histograms represent the grey level distribution of an image. We used
it directly as a feature, as in [7–9], but we also computed some statistical descriptors
([10]) such its mean, standard deviation and skewness (asymmetry). Different functions
can be used to estimate the divergence between two histograms, influencing the quality
of the results.
Texture Features: We tested 6 of the 14 Haralick’s descriptors: contrast, dissimilarity,
second angular moment, mean, homogeneity, entropy, maximal probability and stan-
dard deviation. These are statistical values extracted from the Grey-Level Co-occurrence
55

Matrix (GLCM) of an image, which represents the spatial relationship of a given (dis-
tance, angle) couple, between pixel values. This method is described by Haralick in [11]
and used in numerous CBIR systems, such as [10, 12, 13]. A GLCM in 4 directions is
computed on each block after dividing the image in numerous little windows. We tested
different distances and window sizes.
Shape Features: We tested five shape features: Fourier Descriptors, Procustes analy-
sis and three simple geometric descriptors. After a Fourier transformation, we take as
features the first low frequency normalized coefficients, which are named the Fourier
Descriptors [14]. Many simple geometric measures can be computer over the shape (see
[14, 15]). We chose three of them: circularity (distance from the object shape to an ideal
shape, a circle), eccentricity (principal axis ratio), and variance. Procustes analysis ([14,
16]) determines the best linear transformation between two shapes (translation, rotation,
scale) then returns a distance measure used as a descriptor.
Direct Comparison: Direct comparison criteria cannot be called visual features, but
they can be considered as distance measure functions between the whole images con-
sidered as probability distributions. We tested both linear correlation ([17]) and mutual
information. For two images I1 and I2:
linear correlation =
covariance(I1, I2)
p
covariance(I1, I1) ∗ covariance(I2, I2)
mutual inf ormation =
X
i,j
P (i, j) ∗ log2
P (i, j)
P
1
(i, j) ∗ P
2
(i, j)
with, for each pixel (i,j), P the joint probability distribution function of I1 and I2, and
P
1
and P
2
the marginal probability distribution functions of I1 and I2 respectively.
2.5 Proposed Combination Method
With the use of only one feature, results are not always stable, as illustrated by Fig. 4.
In order to improve individual results, the informations from several features are com-
bined. Often, each feature is given a weight and the results are computed accordingly
to these weights. Weights can be determined either in an arbiratry way (all features are
of equal importance, [18]) or by relevance feedback or with a learning algorithm [19].
We present a different method, which reduces computational time, based on successive
refinements of the results. Its framework is shown on Fig. 5. We applied this algorithm
on the three features retrieving the results of best quality on our dataset. The best tenth
results given by the comparison of the first individual feature are returned. It determines
a low and high limits of search in our reference dataset, i.e. in the body range. We then
compute the second feature on the reference database in the images within these bound-
aries. Finally, the third feature is compared to the images in the range determined by the
second feature in the reference database. This method allows the distance measurement
between the test feature and a limited number of the reference dataset descriptors.
56
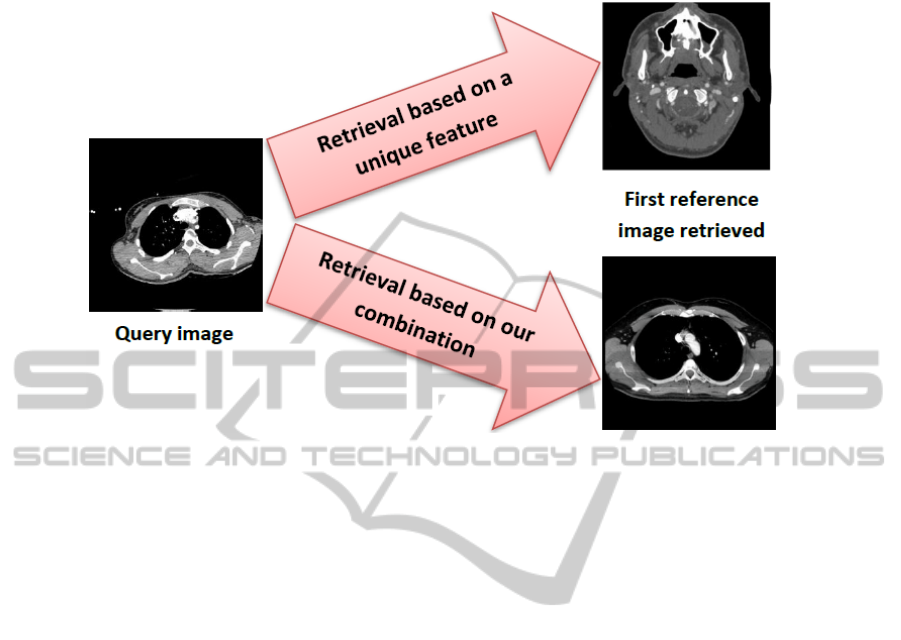
Fig. 4. Combination process.
3 Results and Discussion
We chose, in order to evaluate our method, two criteria. The first one is the graph Pre-
cision versus Recall. The second one is the distance in millimiters between the ideal
image and the image retrieved. A radiologist determined for each test image the closest
image in the reference database. Our results are presented in this section.
3.1 Distance
Given that each image in the reference database is 3 millimeters distant to its neigh-
bours, we computed the distance between the top ten images retrieved and the ideal
image. Then, cumulative distance was calculated for each test image and its ten first re-
sults, as well as the total cumulative distance for the whole dataset. These values will be
used to make comparisons in our future work. We obtain 48mm as an average distance
for the first image returned to each of the ten test images, which we estimate as a good
start for our system.
Some results and their associated distances are presented in Figure 6.We can see
that results are stable: for a query image, most retrieved images are very close between
them. In shown results, the closest reference image is retrieved in second, fifth and third
position from top to bottom query. In future work, we plan to improve these positions.
3.2 Precision vs. Recall
We decided to follow the idea proposed by [20]. We determined a pertinence criterion
based on the distance defined above: a retrieved image is estimated pertinent when its
57
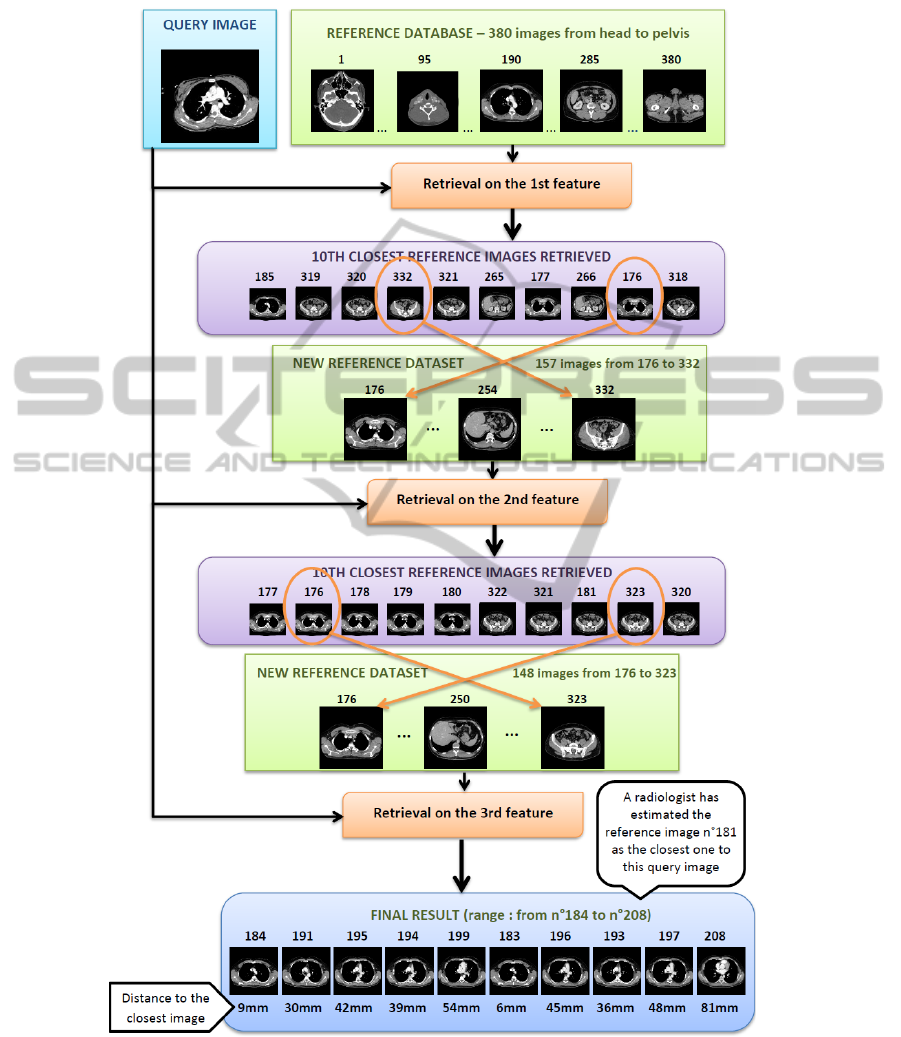
Fig. 5. Combination process.
58
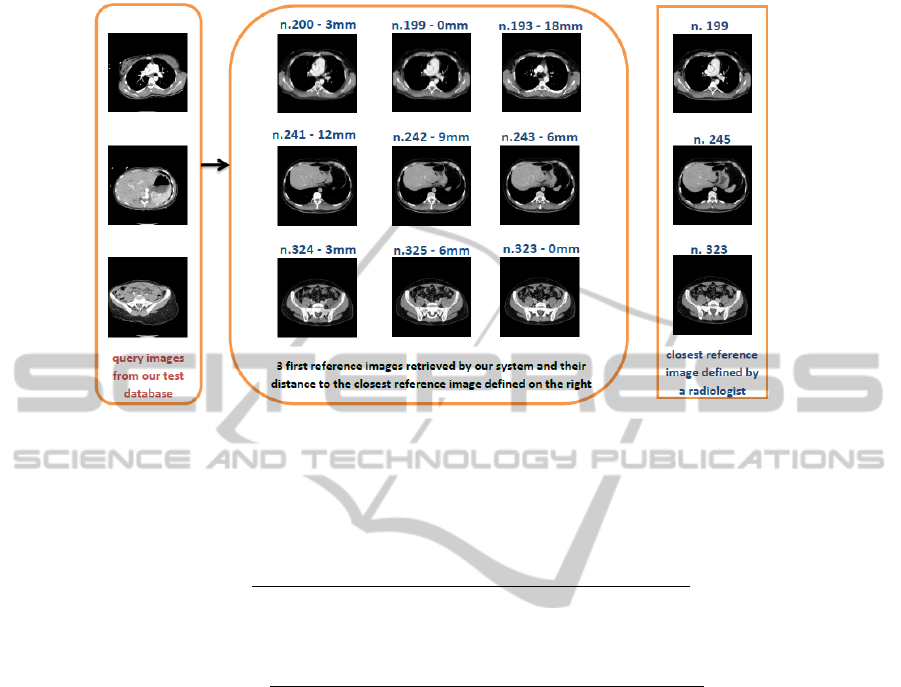
Fig. 6. Distance results - 10 first images retrieved to each query image.
distance to the closest reference image is less or equal to 30mm. A radiologist has
estimated this value as a good estimator for our current positioning system. For each
pertinent image retrieved, we obtain a recall value, defined as
recall =
number of pertinent results retrieved until here
total number of pertinent results in the database
as well as an associated value of
precision =
number of pertinent results retrieved until here
number of results retrieved unti lhere
The precision can be seen as the system ability to retrieve, in the first results, mostly
pertinent images. The recall, as the capacity of the system to retrieve all pertinent results
of the database.
The mean of the precision values obtained for each recall value is computed for
all the test images and presented Fig 7. The curves show that our combination gives
an overall higher precision than individual features. However, these individual charac-
teristics perform a better precision for some recall values (1/21, 2/21, 3/21 and 8/21).
We intend to work on this point in future work. An ideal combination would not lose
precision in comparison to individual visual features.
4 Conclusions
This paper presents a CBIR system that incorporates a new features combination method
and is dedicated to work on generic CT images. The advantages of our three-steps com-
bination are its multiscale approach, which permits fast retrieval, and its modularity.
59
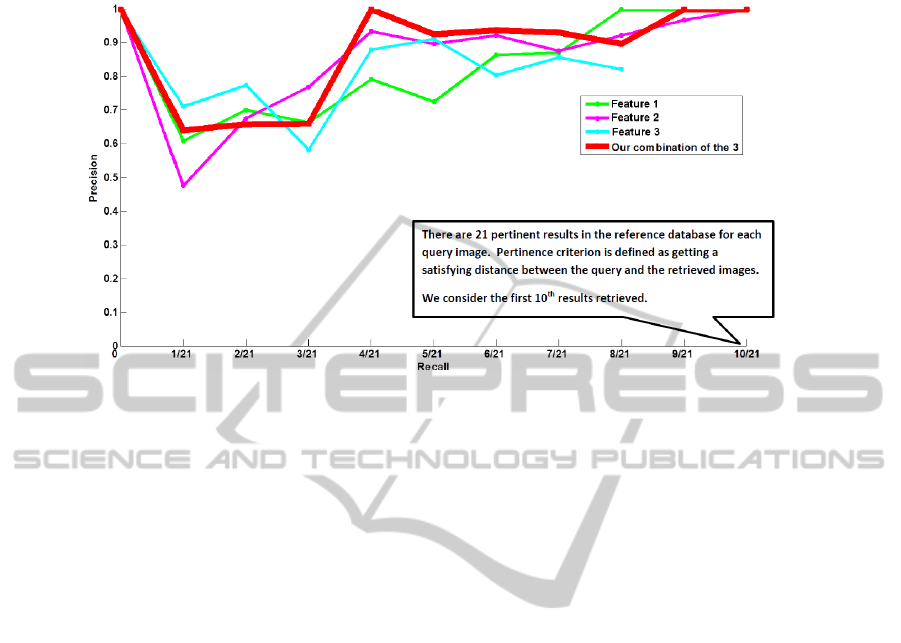
Fig. 7. Mean of Precision values for each Recall value.
This last characteristic will make our next improvements easy to integrate: the number
of steps can be set higher, and features can be permuted or changed. The first results
seem promising. Future work will be to confirm the interest of our method on a larger
test dataset, and compare it to other combination processes.
References
1. Jlia E. E. de Oliveiraa, Alexei M. C. Machadob, Guillermo C. Chaveza, Ana Paula B. Lopesa,
Thomas M. Desernoc, Arnaldo de A. Arajoa, MammoSys: A content-based image retrieval
system using breast density patterns, Computer Methods and Programs in Biomedicine,
2010, Article In Press.
2. William Hsu, Sameer Antani, L. Rodney Long, Leif Neve, George R. Thoma, SPIRS: A
Web-based image retrieval system for large biomedical databases, International Journal of
Medical Informatic 78S, S13-S24, 2009.
3. Thomas M. Deserno, Sameer Antani, Rodney Long, Ontology of Gaps in Content-Based
Image Retrieval, Journal of Digital Imaging, Vol. 22 N. 2, p202-215, 2009.
4. Aur
´
eline Quatrehomme, Deis Hoa, G
´
erard Subsol, William Puech, Review of Features Used
in Content-Based Radiology Image Retrieval systems, International Workshop on Image
Analysis, Nmes (France), August 2010.
5. L. Rodney Long, Sameer Antani, Thomas M. Deserno, George R. Thoma, Content-Based
Image Retrieval in Medicine: Retrospective Assessment, State of the Art and Future Direc-
tions, International Journal of Healthcare Information Systems and Informatics, Vol. 4 n1,
2008.
6. Ceyhun Burak Akgl, Daniel L. Rubin, Sandy Napel, Christopher F. Beaulieu, Hayit
Greenspan, Burak Acar, Content- Based Image Retrieval in Radiology: Current Status and
Future Directions, Journal Of Digital Imaging, 2009.
7. Pedro H. Bugatti, Marcelo Ponciano-Silva, Agma J. M. Traina, Caetano Traina Jr., Paulo M.
A. Marques, Content-Based Retrieval of Medical Images: from Context to Perception, 22nd
IEEE International Symposium on Computer-Based Medical Systems, 2009.
60

8. Juan C. Caicedo, Fabio A. Gonzalez, Eduardo Romero, C. Peters et al., Content-Based Med-
ical Image Retrieval Using Low-Level Visual Features and Modality Identification, CLEF
2007, LNCS 5152, p.615-622, 2008.
9. Edwin A. Nino, Juan C. Caicedo, Fabio A. Gonzales, Metric Indexing for Content-Based
Medical Image Retrieval, Avances en sistemas e Informtica 5 (1), 73-79, 2008.
10. Marcela X. Ribeiro, Pero H.Bugatti, Caetano Traina Jr., Paulo M.A. Marques, Natalia A.
Rosa, Agma J.M. Traina, Supporting content-based image retrieval and computer-aided di-
agnosis systems with association rule-based techniques, Data and Knowledge Engineering
68, p.1370-1382, 2009.
11. Robert M. Haralick, Statistical and Structural Approaches to Texture, Proceedings of the
IEEE Vol.67 n 5, 1979.
12. M.Rahman, B.C.Desai, P.Bhattacharya, Medical image retrieval with probabilistic multi-
class support vector machine classifiers and adaptive similarity fusion, Computerized Medi-
cal Imaging and Graphics 32 (2), 95-108, 2008.
13. A. Mueen, R. Zainuddin, M. Sapiyan Baba, Automatic Multilevel Medical Image Annotation
and Retrieval, Journal of Digital Imaging, 2007.
14. Xiaoqian Xu, Dah-JyeLee, Sameer K. Antani, L. Rodney Long, James K. Archibald, Using
relevance feedback with short-term memory for content-based spine X-ray image retrieval,
Neurocomputing 72, p.2259-2269, 2009.
15. Liyang Wei, Yongyi Yang, Robert M.Nishikawa, Microcalcification classification assisted by
content-based image retrieval for breast cancer diagnosis, Pattern Recognition 42, p.1126-
1132, 2009.
16. Xiaoning Qian, Hemant D. Tagare, Robert K. Fulbright, Rodney Long, Sameer Antani, Op-
timal embedding for shape indexing in medical image databases, Medical Image Analysis
14, p.243-254, 2010.
17. Thomas M. Deserno, Mark O. Gld, Bartosz Plodowski, Klaus Spitzer, Berthold B. Wein,
Henning Schubert, Hermann Ney, Thomas Seidl, Extended Query Refinement for Medical
Image Retrieval, Journal of Digital Imaging 68, 2009.
18. Manesh Kokare, CT Image Retrieval Using Tree Structured Cosine Modulated Wavelet
Transform, International Conference on Digital Image Processing, 2009.
19. Dah-Jye Lee, Sameer Antani, Yuchou Chang, Kent Gledhill, L. Rodney Long, Paul Chris-
tensen, CBIR of spine X-ray images on inter-vertebral disc space and shape profiles using
feature ranking and voting consensus, Data and Knowledge Engineering 68, p.1359-1369,
2009.
20. Sandy A. Napel, Christopher F. Beaulieu, Cesar Rodriguez, Jingyu Cui, Jiajing Xu, Ankit
Gupta, Daniel Korenblum, Hayit Greenspan, Yongjun Ma, Daniel L. Rubin, Automated Re-
trieval of CT Images of Liver Lesions on the Basis of Image Similarity: Method and Prelim-
inary Results, Radiology, Vol. 256 N.1, July 2010.
61
