
A PDMS BASED INTEGRATED PCR MICROCHIP FOR
GENETIC ANALYSIS
Sandeep Kumar Jha, You-Cheol Jang, Rohit Chand, Kamrul Islam
Department of Nanoscience & Engineering, Myongji University, Yongin, 449728, Republic of Korea
Yong-Sang Kim
Department of Nanoscience & Engineering, Myongji University & Department of Electrical Engineering, Myongji
University, Yongin, 449728, Republic of Korea
Keywords: PDMS, PCR Microchip, SMAD4, Lab-on-a-chip, ITO microheater.
Abstract: An integrated continuous-flow microfluidic chip was fabricated on glass substrate with PDMS based
microchannels, cell lysis and PCR modules. Gold-microelectrodes were used to produce electrochemical
cell lysis, while, indium-tin-oxide (ITO) microheater was used for thermal cycling of PCR reaction. The
fabricated device was used for 20 cycles of PCR amplification of pancreatic cancer DNA marker (SMAD4)
from non-tumorigenic MCF10a human cell lines. The 193 bp PCR amplicon obtained through on-chip PCR
was confirmed in case of MCF 10a cells through agarose gel electrophoresis, whereas no product was
detected in case of tumorigenic MCF7 cells. The total time required for entire reaction was less than 90 min.
Therefore, we propose that such microchip can be helpful in predicting the risk of cancer by analysis of
genetic tumor markers from human samples and can also be used for other genetic analysis involving PCR
reaction.
1 INTRODUCTION
Early detection of metastasis has always remained as
elusive as necessity. Such detection often involves
invasive tissue biopsy or expensive and unreliable
tumor marker antigen study. However, the probable
occurrence of a certain forms of cancer can be
predicted as early as gastrulation using genetic
markers. SMAD4 is one such gene, which is either
deleted or mutated in more than one third of
pancreatic cancer patients (Dixit and Juliano, 2008).
These genetic markers can be identified by using
Polymerase chain reaction (PCR), which is a widely
used molecular biology technique for amplifying
specific regions of DNA using DNA polymerase
enzyme. The PCR technique is also applicable in
cloning, genotyping, drugs discovery, forensic,
environmental and ever growing application areas. A
majority of PCR applications involves analysis with
whole cells and requires the extraction of template
DNA prior to PCR. These steps are time consuming
and labor extensive and require expensive chemicals
and instrumentation.
Since, further miniaturization of this technique
is possible, we propose in this regard, a continuous-
flow PCR chip on glass substrate using indium-tin-
oxide (ITO) heater and microchannels laid in
polydimethylsiloxane (PDMS). As, cell lysis is an
important step for extraction of DNA prior to PCR
analysis, for this reason, we also integrated a cell
lysis device on the same chip. While, PDMS was
chosen for microfluidic devices fabrication as it can
easily and repeatedly be fabricated by the molding
method and requires no channel pretreatment as in
case of materials such as Poly(methyl methacrylate)
(PMMA); ITO heater electrodes were the choice for
thermal cycling due to ease of its fabrication and
linear variation of its temperature by application of
DC power.
2 EXPERIMENTAL
The Micro chip consisted of two parts (Fig. 1). The
273
Kumar Jha S., Jang Y., Chand R., Islam K. and Kim Y..
A PDMS BASED INTEGRATED PCR MICROCHIP FOR GENETIC ANALYSIS.
DOI: 10.5220/0003288202730275
In Proceedings of the International Conference on Biomedical Electronics and Devices (BIODEVICES-2011), pages 273-275
ISBN: 978-989-8425-37-9
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)
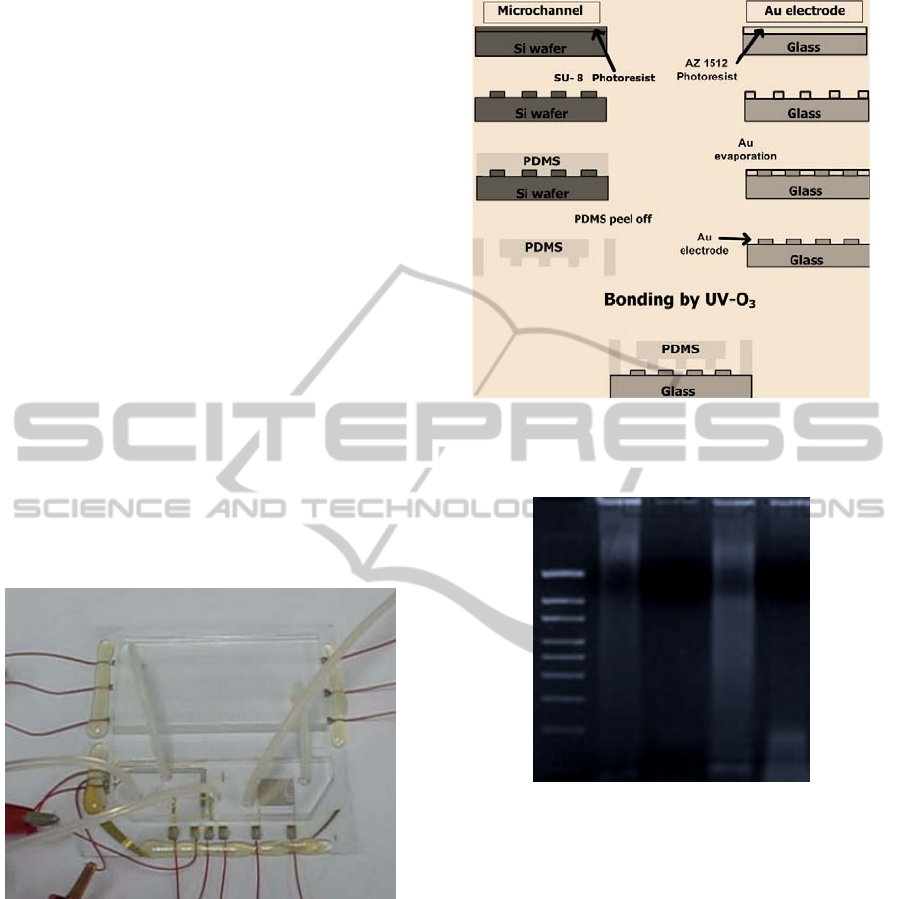
PDMS microchannel was fabricated using negative
molding method. Negative photoresist (SU-8 2075,
Micro Chem) was spin-coated onto a silicon wafer.
SU-8 was patterned to make a microchannel using
photolithography technique (Fig. 2). The PDMS
(DC-184, Dow Corning) mixture was poured on the
SU-8 patterned wafer and cured for 4 h at 72°C. The
PDMS was then peeled off and manual drilling was
performed to produce access holes. The width and
depth of the microchannel were 250 and 200 µm
respectively, and total length was 1100 mm for 20
PCR cycles. The PCR channel was divided into
2:2:3 length ratio for three different temperature
zones namely denaturation, annealing, and
extension. ITO heater/electrode for PCR was
fabricated using conventional photolithography and
wet etch process. Positive photoresist (AZ-1512,
Chariant) was spin-coated on ITO film deposited
glass. AZ-1512 was patterned to make electrode
using photolithography. ITO film was etched using
FeCl
3
/HCl solution for 1 h and photoresist was
removed. Gold electrodes for cell lysis were
fabricated using photolithography and evaporation
method. AZ-1512 was spin-coated on glass and
patterned using photolithography. After
photolithography process, Gold electrode was
Figure 1: The PCR microdevice containing microchannels
for cell lysis and PCR modules, ITO microheater and the
gold microelectrode for electrochemical cell lysis.
deposited using evaporator. For electrical isolation
of electrodes from test fluid, PDMS was spin coated
onto the ITO patterned glass and baked at 95°C for
30 min. Fabricated PDMS microchannel and
ITO/Gold electrode chip were bonded with each
other after UV-ozone treatment for 40 min. ITO
heaters were calibrated for liquid temperature
control by incorporating thermocouple (CHAL-
0001, Omega) into the microchannel during UV-
ozone bonding of PDMS to the glass substrate.
Figure 2: Fabrication process for PDMS based
microchannel and Au microelectrodes.
Lane-1 2 3 4 5
Figure 3: Gel-doc from agarose gel electrophoresis of PCR
amplicon of MCF 7 and MCF 10a cells (SMAD4 gene.
Condition: injecting sample with flow of air at 5 µl/min
rate. Lane: 1= 1.5 kb marker; 2 = Pre PCR lysate for
MCF7; 3 = PCR of MCF7; 4 = Pre PCR lysate for
MCF10a (Non-cancerous); 5 = PCR of MCF10a. The 193
base pair band on lane 5 is the desired PCR product after
20 cycles.
3 RESULTS
The fabricated device was used for lysis and PCR
amplification of genomic DNA of non-tumorigenic
MCF10a and tumorigenic MCF7 human cell lines.
Approximately 10
6
cells were suspended in 50 µl
PBS and 50 µl 2X PCR mix containing Taq-DNA
polymerase and 2 µl each of the primers against
SMAD4 gene were added. The mixture was injected
into silicone tube carrying air to microchannel with
BIODEVICES 2011 - International Conference on Biomedical Electronics and Devices
274

the help of precision syringe pump. The PCR
product (193 bp) was verified by agarose gel
electrophoresis (Fig. 3) as well as spectroscopic
method and a yield of ~ 250 ng DNA/10
6
MCF 10a
cells was recorded, while no product was detected in
case of MCF7. These results suggested the success
of miniaturized PCR device for rapid PCR
amplification of SMAD4 gene in tumorigenic human
cells and therefore, early prediction of occurrence of
pancreatic cancer. In conclusion, the developed
device can also be used in almost any other genetic
analysis involving DNA extraction and PCR
amplification. Based on these preliminary results,
we propose that the integrated device will be helpful
in reducing the reaction time for DNA extraction as
well as PCR amplification of DNA in a variety of
samples. Further work is under progress to use this
integrated chip in genomic analysis from a variety of
samples.
ACKNOWLEDGEMENTS
This study was supported by grant No. ROA-2006-
000-10274-0 from the National Research Laboratory
Program of the Korea Science & Engineering
Foundation.
REFERENCES
Dixit, V. and R. L. Juliano, 2008. Selective killing of
SMAD4-negative tumor cells via a designed repressor
strategy. Mol Pharmacol. 74(1) 289-97.
Joung, S-R., Kang,
C-J. and Kim Y-S, 2008. Series DNA
Amplification Using the Continuous-flow Polymerase
Chain Reaction Chip. Japanese Journal of Applied
Physics. 47(2) 1342-1345.
A PDMS BASED INTEGRATED PCR MICROCHIP FOR GENETIC ANALYSIS
275
