
LEUKOCYTES CLASSIFICATION USING BAYESIAN NETWORKS
Ver´onica Rodr´ıguez-L´opez and Ra´ul Cruz-Barbosa
Computer Science Institute, Universidad Tecnol´ogica de la Mixteca, 69000, Huajuapan, Oaxaca, M´exico
Keywords:
Bayesian networks, Classification, Leukocyte recognition.
Abstract:
In this paper, the use of bayesian networks in the leukocytes classification problem is explored. The com-
plexity in this problem is mainly due to morphological diversity between cells of the same type and similar
features found in different types of cells, which complicate the classification task. Since bayesian networks
have demonstrated to be useful as both a classifier and a powerful tool for knowledge representation and infer-
ence under conditions of uncertainty, this graphical model is applied in the leukocytes classification problem.
The design of two bayesian network models based on the expert’s knowledge and data are presented. Some
preliminary results have shown that the proposed models classify all types of leukocytes with an acceptable
accuracy.
1 INTRODUCTION
White blood cells, or leukocytes, are cells of the
immune system involved in defending the body
against infection. There are five types of leuko-
cytes that normally appear in blood: neutrophils,
basophils, eosinophils, lymphocytes and monocytes
(Greer et al., 2009).
One of the most frequently requested test in a
hematology laboratory is a complete blood count
(CBC). As part of the CBC, a white blood cell count
and a differential white blood cell count are done. The
former measures the total number of white blood cells
in a volume of blood given. The latter consists of
a blood examination to determine the presence and
the number of different types of white blood cells
(Estridge et al., 1999; Carr and Rodak, 2004).
Leukocytes can be counted by either manual or
automated hematology analyzers. The manual leuko-
cytes count is a time consuming task, and highly de-
pendent on lab technician skills who performs the dif-
ferential analysis. Human classification errors are the
main source of misclassification in the manual counts,
where the main problem is the scarcity of cell sam-
ples (usually, sample sizes range from 100 to 200).
On the other hand, automated hematology analyzers
classify cell populations using both electrical and op-
tical techniques. These machines decrease the time of
performing routine examinations and at the same time
increase cells classification accuracy. However, these
analyzers are unable to accurately identify and clas-
sify all types of cells and are, particularly, insensitive
to abnormal or immature cells. For this reason, most
tests performed by these equipments will require a re-
view of a skilled lab technician for cell type definitive
identification (Greer et al., 2009).
To help lab technicians on leukocytes identifica-
tion, many computational systems based on digital
image processing and pattern recognition techniques
have been developed. Despite several systems have
reported a good performance (Colunga et al., 2009;
Mircic and Jorgovanovic, 2006; Rodrigues et al.,
2008), automation of leukocytes recognition is not an
easy task. There are two main problems in this pro-
cess. Firstly, cell morphology is very diverse between
cells of the same type (e.g. neutrophil morphology).
Secondly, different types of cells share some charac-
teristics as shape and texture.
In this work, taking into consideration that the
leukocytes classification is an expert and uncertain
task domain, we explored the use of bayesian net-
works for discrimination of five types of leukocytes.
Bayesian networks have demonstrated to be useful as
both a classifier and a powerful tool for knowledge
representation and inference under conditions of un-
certainty.
This paper is organized as follows. In section 2,
a brief description about bayesian networks is pre-
sented. The description of the bayesian network mod-
els design for leukocytes classification and the corre-
sponding results are presented in section 3. Finally,
some preliminary conclusions are presented in section
4.
681
Rodríguez-López V. and Cruz-Barbosa R..
LEUKOCYTES CLASSIFICATION USING BAYESIAN NETWORKS.
DOI: 10.5220/0003197706810684
In Proceedings of the 3rd International Conference on Agents and Artificial Intelligence (ICAART-2011), pages 681-684
ISBN: 978-989-8425-40-9
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

2 BAYESIAN NETWORKS
Bayesian networks (BN), also known as belief net-
works, belong to the probabilistic graphical models
family. These graphical structures are used for knowl-
edge representation of uncertain domains and when
they work with statistical techniques together, they
present several advantages for data analysis (Hecker-
man, 1996).
A formal definition of a BN is as follows. A
bayesian network model, or simply a bayesian net-
work, is a pair (D, P), where D is a directed acyclic
graph (DAG), P = {p(x
1
|π
1
), ..., p(x
n
|π
n
)} is a set of
n conditional probability distributions, one for each
variable, and Π
i
is the set of parents of node X
i
in D
(Castillo et al., 1997). The set P defines the associated
joint probability distribution as
p(x
1
, x
2
, ..., x
n
) =
n
∏
i=1
p(x
i
|π
i
) (1)
The construction of a bayesian network involves
the definition of its structure and the estimation of its
parameters. In the simplest case, the structure of a
bayesian network is specified by an expert and then
the corresponding parameters are learned from the
available data.
3 EXPERIMENTS
3.1 Experimental Design and Settings
In order to explore the performance of bayesian net-
works in the leukocytes classification problem, we de-
signed two models of this approach. That is, two ex-
periments for classifying all types (neutrophils, ba-
sophils, eosinophils, lymphocytes and monocytes) of
leukocytes were conducted. In the first experiment,
a bayesian network which includes some important
morphological features for leukocytes classification
was built. In the second experiment, we searched for
a simpler bayesian network with a better performance
than the one designed in the first experiment.
For the first experiment, the leukoA model was
developed. In the leukoA model, we proposed a
leukocyte classification node as the main one, and
with the purpose of expressing the real dependence
among features of leukocytes, we used a tree struc-
ture. In this model, we aimed to use some character-
istics that experts take into account for the classifica-
tion process. These features were incorporated into
the model as discrete latent variables. Furthermore,
for the bayesian network structure building we placed
some observable nodes (which are linked to the la-
tent variables) representing the description or mea-
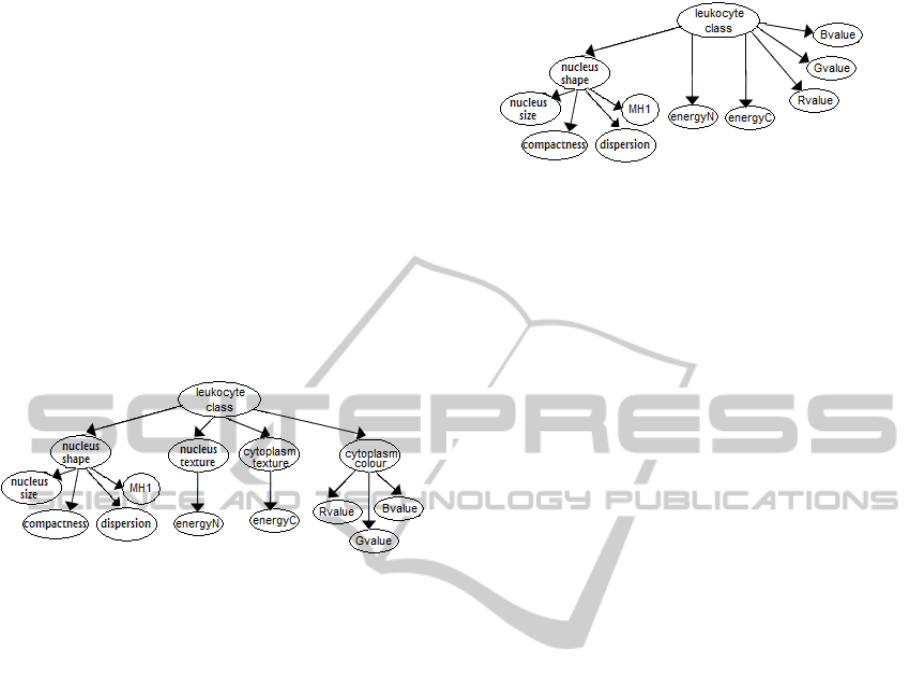
surements of the corresponding features (see Figure
1). These measurements were obtained by applica-
tion of digital image processing techniques. The ob-
servable nodes are continuous variables that have a
normal distribution. The description of the incorpo-
rated knowledge into the leukoA model is presented
as follows.
The first characteristic considered into the leukoA
model was the shape of the nucleus. The nucleus
shape of lymphocytes is round, and the monocytes
shape have a great reniform or horseshoe-shaped nu-
cleus. The nucleus of neutrophils have from 2 to 5
lobules, it can present S, C or glass shapes. The nu-
cleus of eosinophils have 2 lobules and usually it is
glass shaped. The nucleus of basophils is bi- or tri-
lobed, but it is hard to see because of the number of
granules which hide it (Carr and Rodak, 2004; Greer
et al., 2009; Estridge et al., 1999). This knowledge
about the shape of nucleus was encoded into the nu-
cleus shape node. The estimation of this shape was
obtained by means of region descriptors, particularly,
we used the compactness, dispersion and the first Hu
moment (Nixon and Aguado, 2007). These descrip-
tors were included into the leukoA model as compact-
ness, dispersion and MH1 nodes.
Since nucleus size is more relevantthan cytoplasm
size for leukocytes identification, only the nucleus
size was considered for the leukoA model. For the
nucleus size measurement we took the number of pix-
els that belong to the corresponding region divided by
the total number of pixels of the cell (nucleus and cy-
toplasm pixels). This nucleus size information was
included into the nucleus size node, which was linked
with the nucleus shape node due to the relationship
between these two features.
The cytoplasm texture is an important characteris-
tic of leukocytes, it allows to group the cells by the
presence or absence of granules in their cytoplasm
(Greer et al., 2009). The granulocyte type cells are
neutrophils, basophils and eosinophils. The agranu-
locyte cells are lymphocytes and monocytes. In order
to get information about the cytoplasm texture, the en-
ergy descriptor (Nixon and Aguado, 2007) was used.
This knowledge about the cytoplasm texture and its
corresponding descriptor were captured with the cy-
toplasm texture and energyC nodes.
The texture of nucleus is another important char-
acteristic of leukocytes that is reported in medical
literature (Greer et al., 2009; Estridge et al., 1999).
For this reason, we included this knowledge into the
leukoA model in a similar way as the cytoplasm tex-
ture was.
ICAART 2011 - 3rd International Conference on Agents and Artificial Intelligence
682
The colour of cytoplasm was the last feature of
leukocytes taken into account for the leukoA model.
The granulocyte leukocytes are characterized by the
presence of differently staining colour granules in
their cytoplasm: neutrophils have pink colour gran-
ules, eosinophils have orange granules, and basophils
have dark purple granules. For the agranulocyte
cases, the cytoplasm colour for lymphocytes is light
blue and for monocytes is greyish blue (Carr and Ro-
dak, 2004; Estridge et al., 1999). The colour de-
scriptor was obtained through the average intensity
value using the RGB space. The knowledge about
the colour was encoded into the cytoplasm colour,
Rvalue, Gvalue and Bvalue nodes.
In summary, the topology of the LeukoA model is
showed in Figure 1.
Figure 1: Topology of the leukoA bayesian network model
for leukocytes classification.
For the second experiment, we explored the pos-
sibility to find a tree type bayesian network model
with a minimum set of nodes, that performs leuko-
cyte classification with an acceptable degree of accu-
racy. A definition of the new model was found by
modifying the leukoA model. The modification is as
follows. Analyzing the leukoA model, we observed
that the cytoplasm colour node is a redundant node
because it does not encode uncertain information. For
this reason, the cytoplasm colour node was removed.
Since either cytoplasm or nucleus texture is described
by one measurement we decided to remove the cyto-
plasm and nucleus texture nodes in the leukoA model.
We hypothesize that the energy’s nodes are enough to
consider the texture information. Following the pre-
vious observations, we defined the second bayesian
network model, named leukoB. The topology of the
leukoB model is presented in Figure 2.
3.2 Preliminary Results
In order to evaluate the performance of the leukoA
and leukoB bayesian network models, we used a set
of 190 leukocytes colour images with a resolution of
256X256 pixels. The images were obtained with the
help of a microscope that has an in-built CCD cam-
Figure 2: Topology of the leukoB bayesian network model
for leukocytes classification.
era with the resolution of 640X480 pixels. The man-
ual selection and cut of leukocytes region were ap-
plied to all images. For the nucleus and cytoplasm
segmentation, we used a free software developed by
Zoltan Kato (Berthod et al., 1996). The image set was
formed by 8 basophils, 72 neutrophils, 9 eosinophils,
31 monocytes and 70 lymphocytes. All images were
previously classified by a human expert.
The classification performance of the designed
bayesian networks was evaluated by five-fold cross-
validation. The parameters of the corresponding
models were obtained by using maximum likeli-
hood estimation from complete data (Heckerman,
1996). The models were tested using the Hugin Lite
7.3
R
software.
The average classification accuracy results for
the proposed bayesian network models are shown in
Table 1. These results are slightly better for the
leukoB model, which favours the simpler model as
we expected. Also, the results from Table 1 com-
pare favourably with those of alternative methods
that consider less types of leukocytes than the ones
used here. For example, the classifiers presented in
(Mircic and Jorgovanovic, 2006) and (Colunga et al.,
2009) are complex and consider only the most com-
mon types of leukocytes. In (Mircic and Jorgov-
anovic, 2006), they only classify four types (neu-
trophils, eosinophils, lymphocytes and monocytes) of
leukocytes with 86% of accuracy. While in (Colunga
et al., 2009), they classify neutrophils, eosinophils
and lymphocytes with 84% of accuracy. In contrast,
our bayesian network models are simple classifiers
that can be easily understood and verified by experts.
In Table 2, the average classification accuracy re-
sults for each type of leukocyte is presented. From
this table, it can be observed that both models
can classify all types of leukocytes, including ba-
sophils and eosinophils, which are, usually, imbal-
anced classes (they appear less frequently in blood
cells). These preliminary results show that bayesian
networks are promising models for leukocytes classi-
fication.
LEUKOCYTES CLASSIFICATION USING BAYESIAN NETWORKS
683
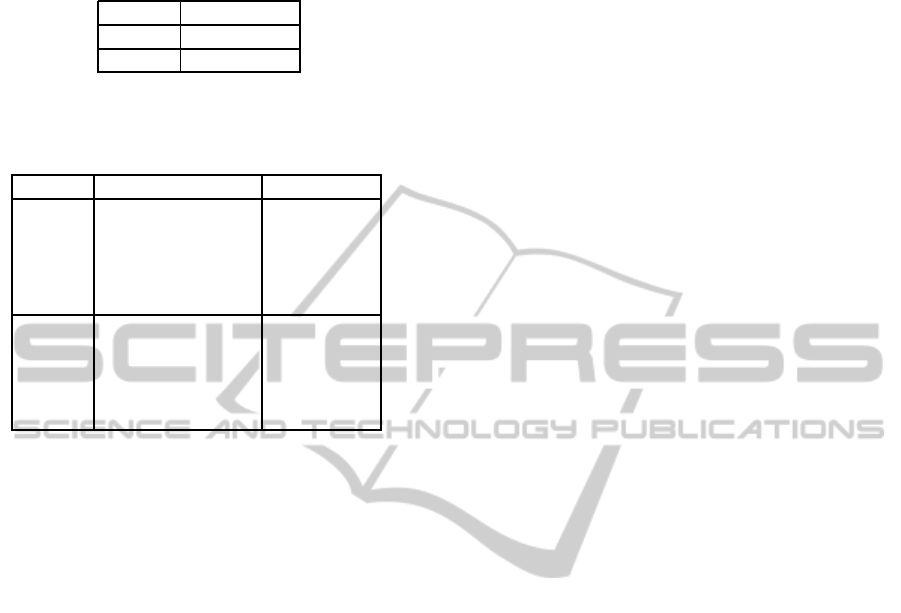
Table 1: Average classification accuracy results (using
cross-validation) for leukoA and leukoB bayesian network
models.
Model classif. acc.
leukoA 87.9%
leukoB 90.5%
Table 2: Average classification accuracy results for each
type of leukocyte of the leukoA and leukoB bayesian net-
work models.
Model type of leukocyte classif. acc.
basophils 93.3%
neutrophils 95.3%
leukoA eosinophils 83.3%
monocytes 61.0%
lymphocytes 88.0%
basophils 93.3%
neutrophils 95.3%
leukoB eosinophils 83.3%
monocytes 82.7%
lymphocytes 89.5%
4 CONCLUSIONS
We presented two bayesian network models for leuko-
cytes classification in this paper. A tree structure
for them and definition of variables by using expert’s
knowledge and medical literature was proposed. De-
spite the analyzed data set have not enough images
of some types of leukocytes (imbalanced classes), the
proposed bayesian networks performance is compa-
rable with those of reported in literature. Our pro-
posed models can classify all types of leukocytes, in-
cluding the less frequent types, with a high degree of
accuracy. These preliminary results have shown that
bayesian network models could be competitive with
other types of classifiers.
As future work, we will use the leukocytes fea-
tures found in this analysis for building a naive bayes
and a neural network model, which can then be com-
pared, in terms of average accuracy, with our pro-
posed bayesian network models.
REFERENCES
Berthod, M., Kato, Z., Yu, S., and Zerubia, J. (1996).
Bayesian image classification using markov random
fields. Image and Vision Computing, (14):285–295.
Carr, J. H. and Rodak, B. F. (2004). Clinical Hematology
Atlas. Saunders, 2nd. edition.
Castillo, E., Gutierrez, J. M., and Hadi, A. S. (1997).
Experts systems and Probabilistic Networks Models.
Springer-Verlag.
Colunga, M. C., Siordia, O. S., and Maybank, S. J.
(2009). Leukocyte recognition using EM-algorithm.
In Aguirre, A. H., Borja, R. M., and Garc´ıa, C.
A. R., editors, MICAI ’09: Proceedings of the 8th
Mexican International Conference on Artificial Intel-
ligence, pages 545–555. Springer-Verlag.
Estridge, B. H., Reynolds, A. P., and Walters, N. J. (1999).
Basic Medical Laboratory Techniques. Delmar Cen-
gage Learning, 4th. edition.
Greer, J. P., Foerster, J., Rodgers, G. M., Paraskevas, F.,
Glader, B., Arber, D. A., and Robert T. Means, J.
(2009). Wintrobe’s Clinical Hematology, volume 1.
Lippincott Williams & Wilkins, 12th. edition.
Heckerman, D. (1996). A tutorial on learning with bayesian
networks. Technical report, Microsoft Research.
Mircic, S. and Jorgovanovic, N. (2006). Automatic classi-
fication of leukocytes. Journal of Automatic Control,
16(1):29–32.
Nixon, M. S. and Aguado, A. S. (2007). Feature Extraction
& Image Processing. Academic Press, 2nd. edition.
Rodrigues, P., Ferreira, M., and Monteiro, J. (2008).
Segmentation and classification of leukocytes using
neural networks: A generalization direction. In
Bhanu Prasad, S. M. P., editor, Speech, Audio, Image
and Biomedical Signal Processing using Neural Net-
works, pages 373–396. Springer Berlin / Heidelberg.
ICAART 2011 - 3rd International Conference on Agents and Artificial Intelligence
684
