
STUDYING THE RELEVANCE OF BREAST IMAGING FEATURES
Pedro Ferreira, Inˆes Dutra
Department of Computer Science & CRACS-INESC Porto LA, University of Porto, Porto, Portugal
Nuno A. Fonseca
CRACS-INESC Porto LA, Porto, Portugal
Ryan Woods
Department of Radiology, Johns Hopkins Hospital, Baltimore, MD, U.S.A.
Elizabeth Burnside
Department of Radiology, University of Wisconsin School of Medicine and Public Health, Madison, WI, U.S.A.
Keywords:
Mass density, Breast cancer, Mammograms, Classification methods, Data mining, Machine learning.
Abstract:
Breast screening is the regular examination of a woman’s breasts to find breast cancer in an initial stage. The
sole exam approved for this purpose is mammography that, despite the existence of more advanced technolo-
gies, is considered the cheapest and most efficient method to detect cancer in a preclinical stage.
We investigate, using machine learning techniques, how attributes obtained from mammographies can relate
to malignancy. In particular, this study focus is on how mass density can influence malignancy from a data
set of 348 patients containing, among other information, results of biopsies. To this end, we applied different
learning algorithms on the data set using the WEKA tools, and performed significance tests on the results. The
conclusions are threefold: (1) automatic classification of a mammography can reach equal or better results than
the ones annotated by specialists, which can help doctors to quickly concentrate on some specific mammogram
for a more thorough study; (2) mass density seems to be a good indicator of malignancy, as previous studies
suggested; (3) we can obtain classifiers that can predict mass density with a quality as good as the specialist
blind to biopsy.
1 INTRODUCTION
Breast screening is the regular examination of a
woman’s breasts to find breast cancer earlier. The sole
exam approved for this purpose is mammography.
Usually, findings are annotated through the Breast
Imaging Reporting and Data System (BIRADS) cre-
ated by the American College of Radiology. The BI-
RADS system determines a standard lexicon to be
used by radiologists when studying each finding. De-
spite the existence of more advanced technologies,
mammography is considered the cheapest and most
efficient method to detect cancer in a preclinical stage.
In this work, we were provided with 348 cases of
patients that went through mammography screening.
Our main objective is to apply machine learning tech-
niques to these data in order to find non trivial rela-
tions among attributes, and learn models that can help
medical doctors to quickly assess mammograms.
Much work has been done on applying ma-
chine learning techniques to the study of breast
cancer, which is one of the most common kinds
of cancer in the world. In the UCI (University
of California, Irvine) machine learning repository
(http://archive.ics.uci.edu/ml/datasets.html) there are
four data sets whose main target of study is breast
cancer. One of the first works on applying machine
learning techniques to breast cancer data dates from
1990. The data set used in this study, donated to the
UCI repository, was created by Wolberg and Man-
gasarian after their work on a multisurface method
of pattern separation for medical diagnosis applied
337
Ferreira P., Dutra I., A. Fonseca N., Woods R. and Burnside E..
STUDYING THE RELEVANCE OF BREAST IMAGING FEATURES.
DOI: 10.5220/0003172903370342
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2011), pages 337-342
ISBN: 978-989-8425-34-8
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

to breast cytology (Wolberg and Mangasarian, 1990).
Most works in the literature applies artificial neural
networks to the problem of diagnosing breast cancer
(e.g., (Wu et al., 1993) and (Abbass, 2002)). Oth-
ers focus on prognostic of the disease using inductive
learning methods (e.g., (Street et al., 1995)). More
recently, Ayer et al. (Ayer et al., 2010) have evalu-
ated whether an artificial neural network, trained on
a large prospectively collected data set of consecutive
mammography findings, could discriminate between
benign and malignant disease, and accurately predict
the probabilityof breast cancer for individual patients.
Other recent studies focus on extracting information
from free textthat appears in medical records of mam-
mography screenings (Nassif et al., 2009), and on the
influence of age in ductal carcinoma in situ (DCIS)
findings (Nassif et al., 2010).
Our study is focused on the influence of mass
density on predicting malignancy, but we also un-
cover other interesting complementary findings. Pre-
vious works by Jackson et al. (Jackson et al., 1991)
and Cory and Linden (Cory and Linden, 1993) have
argued that, although the majority of high density
masses are malignant, the presence of low density
cancers and more important indicators (like margins,
shape, and associated findings) make mass density
a less reliable indicator or predictor of malignancy.
Sickles (Sickles, 1991) has the same opinion. A study
carried out by Davis et al. (Davis et al., 2005) indi-
cated that mass density could have more importance
and relevance than previous works had reported. In
another work, Woods et al. (Woods et al., 2009) ap-
plied inductive logic programming to a set of breast
cancer data and concluded the same thing. Woods and
Burnside (Woods and Burnside, 2010) also applied
logistic regression and kappa statistics to another set
of breast cancer data and concluded that mass density
and malignancy are somewhat related.
In this work, we use the same data set used by
Woods and Burnside (Woods and Burnside, 2010),
but we apply machine learning methods and confirm
the findings of Woods and Burnside. In addition,
we show that the learned classifiers generated in this
work can predict mass density and outcome (classi-
fication of a mammography) with a quality as good
as a specialist, proving to be good helpers to medical
doctors when evaluating mammograms.
2 BREAST CANCER DATA
Our study analyzes 348 consecutive breast masses
that underwent image guided or surgical biopsy per-
formed between October 2005 and December 2007
on 328 female subjects. All 348 biopsy masses were
randomized and assigned to a radiologist blinded
to biopsy results for retrospective assessment us-
ing the Breast Imaging Reporting and Data Sys-
tem (retrospectively-assessed data set). Clinical ra-
diologists prospectively assessed the density of 180
of these masses (prospectively-assessed data set).
Pathology result at biopsy was the study endpoint.
The atributes included in our study are very much
the ones collected by the radiologists from the mam-
mograms, and are based on the BIRADS lexicon.
We selected from the original database all the at-
tributes considered relevant by the specialists and re-
moved some attributes such as identifiers, redundant
attributes and attributes that had the same value for all
instances. For our main task, to predict malignancy,
our class attribute was the outcome binary variable as-
suming values benign or malignant.
From the 348 cases, 118 are malignant (≈ 34%),
and 84 cases have high mass density (≈ 24%) retro-
spectively assessed. Other attributes are mass shape,
mass margins, depth, size, among others. For the
purpose of our study, we have two attributes that
represent the same characteristics of the finding, but
with different interpretations. These are retro density
and density num. Both represent mass densities that
can assume values high or iso/low. Retro density
was retrospectively assessed while density num was
prospectively (at the time of imaging) assessed.
3 EXPERIMENTS AND RESULTS
Our first preliminary study was to calculate simple
frequencies from the data and to determine if there
was some evidence of relationship between attributes,
specially, the main focus of our study:
Is mass density related to malignancy?
As mentioned above, from the 348 breast masses,
118 are malignant (≈ 34%), and 84 have high mass
density (≈ 24%). If we consider that mass density
and malignancy are independent, and take 84 cases
from the 348 at random, the probability of these be-
ing malignant should still be ≈ 34%. However, if it
happens that all 84 cases selected at random have high
density, then the percentage of malignant cases raises
to 70.2% (this is the percentage of cases that are both
malignant and have high mass density). The proba-
bility of this being coincidence is very low, given the
data distribution. This simple calculation may already
imply that high density has some relation to malig-
nancy. So may imply that other attributes such as age,
mass shape and mass margins can have some relation
to malignancy. One of the objectives of our study is
HEALTHINF 2011 - International Conference on Health Informatics
338

then to confirm if these attributes have some relation
to the outcome variable.
3.1 Methods
As mentioned before, the data set used in the experi-
ments contains 348 findings that include data related
to biopsies. A subset of 180 was annotated by a spe-
cialist blind to the biopsies results. The task of this
specialist was to annotate the mass density. The re-
maining findings, 168 cases, were not annotated by
this specialist.
All experiments were performed using the
WEKA tool, developed at Waikato University, New
Zealand (Hall et al., 2009). We experimented with
several classification algorithms, but report only for
the algorithms that produced the best results. The ex-
periments were performed in WEKA using the Exper-
imenter module, where we set several parameters, in-
cluding the statistical significance test and confidence
interval, and the algorithms we wanted to use (we
used OneR as reference, ZeroR, PART, J48, Simple-
Cart, DecisionStump, Random Forests, SMO, Naive
Bayes, Bayes with TAN, NBTree and DTNB). The
WEKA experimenter produces a table with the per-
formance metrics of all algorithms with an indica-
tion of statistical differences, using one of the algo-
rithms as a reference. The significance tests were per-
formed using standard corrected t-test with a signifi-
cance level of 0.01. The parameters used for the learn-
ing algorithms are the WEKA defaults. In the tables,
the numbers between parentheses represent standard
deviations. From the 348 cases, we trained on the 180
annotated cases. We used the remaining 168 as un-
seen/test data to evaluate the performance of the clas-
sifiers. During the training, we used 10-fold stratified
cross validation and reported the results for the aver-
age metrics obtained among all folds.
3.2 Is Mass Density Predictive of
Malignancy?
We considered at least two ways of investigating if
mass density is predictive of malignancy. The first
one is to apply association rules or logistic regression
to the 348 findings, and report the relation between
retro density and outcome. This was already done by
Woods and Burnside (Woods and Burnside, 2010), in
a previous work, using logistic regression and kappa
statistics. Their results showed that high mass density
is a relatively important indicator of malignancy with
an inter-observer agreement of 0.53.
The second way is to use a classification method
and predict outcome using mass density and with-
out using mass density and compare results. As we
have two kinds of mass density: one for the retrospec-
tive data and another one for the prospective data, we
used both to build classifiers. Our first experiment
was then to generate a classifier to predict outcome
with retro density using 10-fold cross-validation on
the 180 findings. Our second experiment was to gen-
erate a classifier to predict outcome with density num
(prospectively assessed), also using 10-fold cross-
validation on the 180 findings.
In order to investigate if mass density is predictive
of malignancy, we also generated a classifier to pre-
dict outcome without any information about density
using 10-fold cross-validation on the 180 findings.
In the three experiments, the best classifiers found
were based on Support Vector Machines (Platt, 1998).
Table 1 summarizes the results obtained using the
metrics we found more relevant to the task. CCI is
the percentage of Correctly Classified Instances. K is
the k-value of kappa statistics. Prec is the Precision,
and F is the F-measure. These results show that mass
density has some influence on the outcome, specially
when mass density is the one observed on the retro-
spective data. The classifier trained without mass den-
sity has an overall performance of 81.39% while the
classifier trained with the retrospective assessed mass
has an overall performance of 84.78%. These results
are statistically different (p=0.01). If we look at the K
value, we can confirm that the relation between mass
density and outcome is not by chance, given the rela-
tively high observed agreement between the real data
and the classifier’s predicted values. With respect to
Precision, the results also seem to be quite good with
only 16% of cases being incorrectly classified as ma-
lignant when using the retrospective data. The Re-
call also gives a reasonable rate of correctly classified
cases of malignancy, although there is still scope for
improvement. The f-measure balances the values of
Precision and Recall and also indicates that the clas-
sifiers are behaving reasonably well.
Summarising, these results show that attributes
other than mass density are also important, but if
we add mass density, the classifier’s performance im-
proves.
These results also confirm findings in the litera-
ture regardingthe relevanceof mass density, and show
that good classifiers can be obtained to predict out-
come (with a high percentage of correctly classified
instances and good values of K, precision and recall).
STUDYING THE RELEVANCE OF BREAST IMAGING FEATURES
339
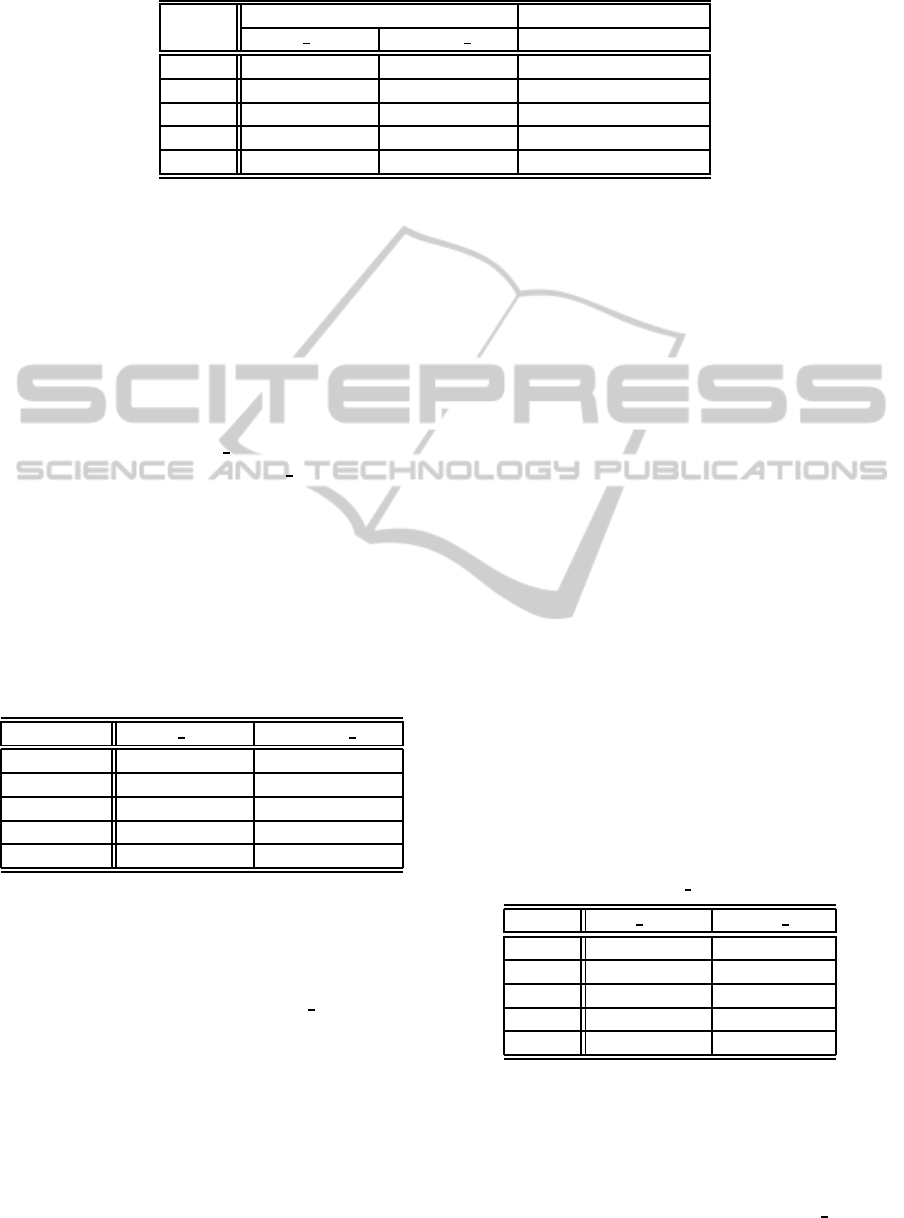
Table 1: Prediction of outcome using 180 findings. Standard deviation values are between parentheses.
Metric with mass density without mass density
retro density density num
CCI 84.78% (7.96) 82.72% (8.32) 81.39% (8.81)
K 0.68 (0.17) 0.63 (0.17) 0.60 (0.18)
Prec 0.84 (0.12) 0.82 (0.13) 0.81 (0.14)
Recall 0.78 (0.15) 0.75 (0.15) 0.72 (0.15)
F 0.80 (0.11) 0.77 (0.11) 0.75 (0.12)
3.3 Can we Obtain a Classifier that
Predicts Mass Density as Well as the
Radiologist?
Our second question is related to the quality of the
classifier related to a specialist. As we have two an-
notated mass densities, one for the prospective study
and another one for the retrospective, we generated
2 classifiers: one is trained on the prospective val-
ues of mass density (density num), and another one
is trained on the retrospective (retro density) values
of mass density. Once more, we used the 180 cases
as training set and 10-fold stratified cross-validation.
The best classifier obtained by the WEKA Exper-
imenter for these two tasks was based on Naive-
Bayes (John and Langley, 1995). Table 2 shows the
results of these experiments as an average of the met-
rics for the 10 folds.
Table 2: Prediction of mass density using 180 findings.
Standard deviation values are between parentheses.
Metric retro density density num
CCI 72.83% (9.89) 67.22% (12.14)
K 0.37 (0.23) 0.33 (0.25)
Precision 0.58 (0.20) 0.66 (0.16)
Recall 0.58 (0.22) 0.60 (0.17)
F-Measure 0.56 (0.18) 0.62 (0.15)
70% of masses annotated by the specialist on the
180 findings agreed to the annotated masses of the
retrospective study. The Naive Bayes classifier pre-
dicted ≈ 73% of correct instances when training on
the retrospective annotated mass (retro density) and
≈ 67% when training on prospective masses anno-
tated by a radiologist. These results are quite good
and indicate that the Bayesian classifier generated in
this study can be well applied as a support tool to
help doctors predicting mass density for unseen mam-
mograms. The values of K, Precision, Recall and f-
measure for this experiment are not so good as the
ones obtained when trying to learn outcome. How-
ever, the K valueindicates that the Naive Bayes classi-
fier has some level of agreement with the actual data,
which is not by chance. One interesting thing to ob-
serve is that, although the classifier trained on the ret-
rospectivedata has a higher rate of correctly classified
instances, it has lower values for Precision, Recall and
f-measure than the classifier trained on the prospec-
tive data. This may indicate that this could be a better
classifier to be used when one does not have informa-
tion about the biopsy data.
Our last question is related to how well a learned
classifier can predict the outcome (malignant or be-
nign) on unseen data blind to the result of the biopsy.
3.4 Can the Generated Classifiers
Behave Well on Unseen Data?
In order to answer this question we need again to
consider classifiers generated using the retrospective
mass density attribute and the prospective mass den-
sity attribute. The first classifier, based on the retro-
spective values of mass density was generated when
training on the 180 findings to answer our first ques-
tion: “is mass density related to malignancy?” This is
a classifier based on Support Vector Machines. How-
ever, we can use yet another classifier, based on the
prospective values of mass density to predict the 168
unseen cases.
Table 3: Prediction of mass density on unseen data.
Metric retro density density num
CCI 82.14% 75.60%
K 0.45 0.35
Prec 0.48 0.38
Recall 0.68 0.71
F 0.56 0.49
As the 168 unseen cases do not have any prospec-
tive annotated mass density, we will fill up these miss-
ing values using the classifiers generated when an-
swering our question 2 (Subsection 3.3). In those
experiments, we generated two classifiers to predict
mass density: one that was trained on retro density
HEALTHINF 2011 - International Conference on Health Informatics
340

Table 4: Prediction of outcome on unseen data.
Metric with mass density w/o mass density
retro density retro density density num
(actual) (fill up by NB) (fill up by NB)
CCI 81.55% 79.76% 79.17% 77.38%
K 0.52 0.48 0.46 0.42
Prec 0.70 0.65 0.65 0.61
Recall 0.60 0.60 0.55 0.53
F 0.64 0.62 0.60 0.57
Table 5: Prediction of mass density.
Metric Mass Density
Radiologist density num density num retro density retro density
(180) (180) (168) (180) (168)
CCI 70.00% 67.22% (12.14) 75.60% 72.83% (9.89) 82.14%
K – 0.33 (0.25) 0.35 0.37 (0.23) 0.45
Prec – 0.66 (0.16) 0.38 0.58 (0.20) 0.48
Recall – 0.60 (0.17) 0.71 0.58 (0.22) 0.68
F – 0.62 (0.15) 0.49 0.56 (0.18) 0.56
and another one that was trained on density num.
Both are Bayesian classifiers. Once we fill up these
values, we can apply a classifier learned to predict
outcome to this unseen data set.
Results of the prediction of mass density on the
unseen data are shown in Table 3. These results were
produced by the best classifier that was, in both cases,
a naive Bayes network.
These results are very good, given that both clas-
sifiers have a prediction performance on the unseen
data well above the one obtained on the training set
(180 cases) with respect to CCI. The K-statistics and
the Recall also improved on the unseen data. We see
a slightly fall in performance when predicting benign
cases, and this is observed by the precision and f-
measure values in the unseen data. The rate of false
positives increases on the unseen data. On the other
hand, the algorithm performs better on classifying the
malignant cases.
Once the predicted values of mass densities of
the 168 findings are filled, we move to the next step,
which is to predict outcome for the unseen data. Re-
sults of this experiment can be found in Table 4.
In Table 4 we show three different predictions
for outcome, using three different sources for the
mass density. The second column in Table 4 shows
the results of predicting outcome using the attribute
for mass density available on the retrospective data
(retro density attribute). The third and fourth columns
show the predictions when using the mass density
filled up by the two Naive Bayes classifiers (one that
was trained on the retro density attribute and another
that was trained on the prospective density num at-
tribute).
Regarding the comparison among these three pre-
dictions we can observe that the three classifiers be-
haved relatively well on the unseen data, capturing
most of the malignant and benign cases. The K value,
once more, indicates that those results are not by
chance. In other words, the classifiers are actually
helping to distinguish between malignant and benign
cases. As observed before, the classifier trained on the
actual retrospective data yields better performance,
but the other classifiers are not performing that far,
which indicates that the lack of biopsy data is not
harming the classification task.
A second observation we take from these results
is that, even using predicted values for mass density
(with prediction errors), the classifiers for outcome
in columns three and four, can maintain a reasonable
performance.
The last conclusion we take from these results is
that mass density is somehow related to outcome, and
is an important attribute that contributes to improve
the performance of the classifiers. A comparison be-
tween the figures on the last column of Table 4 (pre-
diction without mass density) with the figures on the
other columns confirms that fact.
Summarizing, and getting back to our third ques-
tion “3. Can we obtain a classifier that predicts
mass density as well as the radiologist?”, Table 5
shows the performance of all classifiers used for this
task on the training data and on unseen data.
Table 5 summarizes our results for predicting
mass density and shows that the classifiers generated
have a good performance that in some cases is bet-
ter than the one given by the radiologist. The per-
formance on unseen cases is also quite reasonable re-
STUDYING THE RELEVANCE OF BREAST IMAGING FEATURES
341

garding the precision and recall values.
4 CONCLUSIONS AND FUTURE
WORK
In this work, we were provided with 348 cases of
patients that went through mammography screening.
The objective of this work was twofold: i) find non
trivial relations among attributes by applying machine
learning techniques to these data, and; ii) learn mod-
els that could help medical doctors to quickly assess
mammograms. We used the WEKA machine learn-
ing tool and whenever applicable performed statisti-
cal tests of significance on the results.
The conclusions are threefold: (1) automatic clas-
sification of a mammography can reach equal or bet-
ter results than the ones annotated by specialists; (2)
mass density seems to be a good indicator of ma-
lignancy, as previous studies suggested; (3) machine
learning classifiers can predict mass density with a
quality as good as the specialist blind to biopsy.
As future work, we plan to extend this work
to larger data sets, and apply other machine learn-
ing techniques based on statistical relational learning,
since classifiers that fall in this category provide a
good explanation of the predicted outcomes as well
as can consider the relationship among mammograms
of the same patient. We would also like to investi-
gate how other attributes can affect malignancy or are
related to the other attributes.
ACKNOWLEDGEMENTS
This work has been partially supported by the
projects HORUS (PTDC/EIA-EIA/100897/2008)
and Digiscope (PTDC/EIA-CCO/100844/2008)
and by the Fundac¸˜ao para a Ciˆencia e Tecnologia
(FCT/Portugal). Pedro Ferreira has been supported
by an FCT BIC scholarship.
REFERENCES
Abbass, H. A. (2002). An evolutionary artificial neural net-
works approach for breast cancer diagnosis. Artificial
Intelligence in Medicine, 25:265.
Ayer, T., Alagoz, O., Chhatwal, J., Shavlik, J. W., Kahn, C.
E. J., and Burnside, E. S. (2010). Breast cancer risk es-
timation with artificial neural networks revisited: dis-
crimination and calibration. Cancer, 116(14):3310–
3321.
Cory, R. C. and Linden, S. S. (1993). The mammographic
density of breast cancer. AJR Am J Roentgenol,
160:418–419.
Davis, J., Burnside, E. S., Dutra, I. C., Page, D., and Costa,
V. S. (2005). Knowledge discovery from structured
mammography reports using inductive logic program-
ming. In American Medical Informatics Association
2005 Annual Symposium, pages 86–100.
Hall, M., Frank, E., Holmes, G., Pfahringer, B., Reutemann,
P., and Witten, I. H. (2009). The weka data mining
software: An update. SIGKDD Explorations, 11:263–
286.
Jackson, V. P., Dines, K. A., Bassett, L. W., Gold, R. H., and
Reynolds, H. E. (1991). Diagnostic importance of the
radiographic density of noncalcified breast masses:
analysis of 91 lesions. AJR Am J Roentgenol, 157:25–
28.
John, G. H. and Langley, P. (1995). Estimating continuous
distributions in bayesian classifiers. In Proceedings of
the Eleventh Conference on Uncertainty in Artificial
Intelligence, pages 338–345. Morgan Kaufmann, San
Mateo.
Nassif, H., Page, D., Ayvaci, M., Shavlik, J., and Burn-
side, E. S. (2010). Uncovering age-specific invasive
and dcis breast cancer rules using inductive logic pro-
gramming. In Proceedings of 2010 ACM International
Health Informatics Symposium (IHI 2010). ACM Dig-
ital Library.
Nassif, H., Woods, R., Burnside, E., Ayvaci, M., Shavlik,
J., and Page, D. (2009). Information extraction for
clinical data mining: A mammography case study. In
ICDMW ’09: Proceedings of the 2009 IEEE Interna-
tional Conference on Data Mining Workshops, pages
37–42, Washington, DC, USA. IEEE Computer Soci-
ety.
Platt, J. C. (1998). Sequential minimal optimization: A fast
algorithm for training support vector machines. Tech-
nical Report MSR-TR-98-14, Microsoft Research.
Sickles, E. A. (1991). Periodic mammographic follow-up of
probably benign lesions: results in 3,184 consecutive
cases. Radiology, 179:463–468.
Street, W. N., Mangasarian, O. L., and Wolberg, W. H.
(1995). An inductive learning approach to prognos-
tic prediction. In ICML, page 522.
Wolberg, W. H. and Mangasarian, O. L. (1990). Multisur-
face method of pattern separation for medical diagno-
sis applied to breast cytology. In Proceedings of the
National Academy of Sciences, 87, pages 9193–9196.
Woods, R. and Burnside, E. (2010). The mammographic
density of a mass is a significant predictor of breast
cancer. Radiology. to appear.
Woods, R., Oliphant, L., Shinki, K., Page, D., Shavlik, J.,
and Burnside, E. (2009). Validation of results from
knowledge discovery: Mass density as a predictor of
breast cancer. J Digit Imaging, pages 418–419.
Wu, Y., Giger, M. L., Doi, K., Vyborny, C. J., Schmidt,
R. A., and Metz, C. E. (1993). Artificial neural net-
works in mammography: application to decision mak-
ing in the diagnosis of breast cancer. Radiology,
187:81–87.
HEALTHINF 2011 - International Conference on Health Informatics
342
