
A COMPUTATIONAL MODELLING APPROACH TO EXPLORE
THE ANTI-MICROBIAL PRO-DRUG DELIVERY SYSTEM
James T. Murphy, Ray Walshe
Centre for Scientific Computing and Complex Systems Modelling, Dublin City University, Glasnevin, Dublin 9, Ireland
Marc Devocelle
Centre for Synthesis and Chemical Biology, Royal College of Surgeons in Ireland, 123 St. Stephen’s Green, Dublin 2, Ireland
Keywords:
Agent-based model, Antibiotic resistance, Pro-drug, Bacteria, Simulation.
Abstract:
This article documents simulations using an agent-based modelling approach to analyse the system dynamics
of the β-lactamase-dependent therapeutic activation pro-drug delivery system, a novel approach for achieving
selective release of anti-microbial drugs for treating antibiotic-resistant bacteria. It is thought that this strategy
could be a promising approach for treating β-lactamase over-expressing strains of bacteria that are resistant
to traditional β-lactam antibiotics such as penicillin. Test simulations were carried out to investigate the pro-
drug system from a theoretical standpoint and assess the effects of key parameters such as half-life, diffusion
rate and reaction kinetics on the system behaviour. It is important to obtain a thorough understanding of
the complex interplay between the various components involved in the pro-drug delivery system to be able
to interpret results from laboratory testing, and ultimately, from the clinical setting. The agent-based model
described here represents an important stepping stone in connecting the theoretical and practical understanding
of the system as a whole.
1 INTRODUCTION
Our lab is inovolved in developing an agent-based
model, called Micro-Gen, which simulates the growth
of bacterial cells in culture and their interactions
with anti-microbial drug molecules (Murphy et al.,
2007; Murphy et al., 2008; Murphy et al., 2009).
The program uses an agent-based modelling approach
whereby the individual bacterial cells are represented
by unique software agents that are capable of flexi-
ble, autonomous action within a simulated environ-
ment (Jennings et al., 1998). The agent-based ap-
proach means that the system as a whole can exhibit
a complex behaviour that is more than the sum of its
constituent parts. This approach allows the unique dy-
namics within a bacterial colony to be simulated by
taking into account the temporal and spatial hetero-
geneities within the population.
So far, Micro-Gen has been used in previous stud-
ies to examine the role of various low-level cellular
parameters in the response of bacterial populations to
antibiotic treatment (Murphy et al., 2009). Studies
showed that it could accurately predict the minimum
inhibitory concentrations (MIC, a simple laboratory
measure of antibiotic efficacy) for various com-
mon β-lactam antibiotics, including penicillin G and
cephalothin, against methicillin-resistant Staphylo-
coccus aureus (MRSA) bacteria (Murphy et al.,
2008). However, another strength of the model exists
in being able to examine new approaches for treating
antibiotic-resistant bacteria and give insight into po-
tential novel drug treatment strategies. This can aid
in rational drug design by allowing a greater under-
standing of the underlying mechanistic principals de-
termining response to treatment.
In the present study, the existing model has
been adapted to explore a novel approach to treating
antibiotic-resistant bacteria called the β-lactamase-
dependent pro-drug delivery system (Smyth et al.,
2000). This approach involves administering a
substrate-like pro-drug molecule that contains a β-
lactam ring structure (Rautio et al., 2008). Thera-
peutic activation of the pro-drug occurs when its β-
lactam ring structure is cleaved by β-lactamase en-
zymes released from the bacterial cells. This cleavage
results in the selective release of a molecule with anti-
301
T. Murphy J., Walshe R. and Devocelle M..
A COMPUTATIONAL MODELLING APPROACH TO EXPLORE THE ANTI-MICROBIAL PRO-DRUG DELIVERY SYSTEM.
DOI: 10.5220/0003154903010308
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2011), pages 301-308
ISBN: 978-989-8425-36-2
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

microbial properties that kills or inhibits the growth
of the bacterial cells (Stone et al., 2004; Bush et al.,
2004).
This is considered a promising therapeutic ap-
proach because many bacterial species (e.g. Staphylo-
coccus aureus) have evolved to produce β-lactamase
enzymes in response to prolonged clinical exposure
to β-lactam antibiotics such as penicillin G (Abra-
ham and Chain, 1940). The bacterial cells produce β-
lactamase enzymes as a defence mechanism because
the enzymes cleavetheβ-lactamring structure present
in the antibiotic molecules, rendering them inactive
(Fisher et al., 2005). In the USA, it is estimated that
greater than 95% of all S. aureus bacterial isolates
possess resistance to penicillin, due to the expres-
sion of β-lactamase (Levy and Marshall, 2004; Neu,
1992). Therefore, designing pro-drugs that specifi-
cally target these bacteria would introduce an evolu-
tionary selective pressure contrary to that of existing
β-lactam antibiotics.
2 MODEL OVERVIEW
The simulations described here were carried out using
an adapted version of the Micro-Gen Bacterial Sim-
ulator. Previous versions of Micro-Gen were used
to model traditional β-lactam antibiotics. But here
we describe initial efforts to model a new class of
drugs called enzyme-catalysed therapeutic activation
(ECTA) pro-drugs. A detailed description of the un-
derlying program structure along with an analysis of
the mechanistic basis for its output has been previ-
ously published (Murphy et al., 2008; Murphy et al.,
2009).
The individual bacterial cells are represented by
software agents that store physical traits such as their
energy state or amount of antibiotic damage. The
agents also have local behavioural rules associated
with them that dictate their actions during the sim-
ulation. The behaviour of the colony as a whole is
an emergent property of the individual agent interac-
tions. The environment of the simulations is repre-
sented by a discrete, two-dimensional grid contain-
ing diffusible elements such as nutrients, β-lactamase
enzymes and anti-microbial drug compounds (fig. 1).
The movement of these molecules is dictated by a dis-
crete implementation of Fick’s First Law of diffusion
(Ginovart et al., 2002).
The key interaction of the model that determines
the response to pro-drug treatment is the reaction be-
tween the β-lactamase enzyme and the pro-drug. This
reaction is the activation step that triggers the release
of an active drug compound (fig. 2). The success of
Bacterial
cell
-lactamase
enzyme
Antibiotic
molecule
Patch size =
1 !m x 1 !m
Figure 1: Diagram of discrete, two-dimensional simula-
tion environment and key components of simulation (bacte-
rial cells, antibiotic molecules, and β-lactamase enzymes).
Each grid element represents 1 µm
2
area of environment.
the pro-drug approach requires the rapid and specific
release of the active drug compound in the vicinity of
the bacterial cells. The model contains a quantitative
representation of this reaction based on Michaelis-
Menten kinetic theory. The equation for calculating
the reaction rate (V) is as follows:
V =
k
cat
[E]
t
[Ab]
K
M
+ [Ab]
(1)
The key parameters are the turnover rate, k
cat
, and
the Michaelis constant, K
M
. The ratio k
cat
/K
M
is of-
ten used as a measure of enzyme efficiency (Zygmunt
et al., 1992) These parameters can be calculated from
biochemical studies in the laboratory and are specific
to the type of drug used and strain of bacteria. [E]
t
and [Ab] are the concentrations of β-lactamase en-
zyme and antibiotic in the local grid element respec-
tively.
Pro-drug
(inactive)
-lactamase
enzyme
Inside cell
Active anti-microbial
agent
Outside cell
Figure 2: Diagram of activation of pro-drug molecule trig-
gered by cleavage of its β-lactam ring structure by β-
lactamase enzyme. The active component of the pro-drug
can then enter the cell and inhibit growth/destroy the cell
(depending on the mode of action of the drug).
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
302

For the interaction between the activated drug
compound and its target molecule in the bacterial cell
a model of pre-steady state reaction kinetics is used.
The specific target molecule depends on the the type
of anti-microbial drug compound used. It must be
noted that the activated drug is assumed to lack the
β-lactam ring structure, and thus it is not subject to
cleavage and re-inactivation by β-lactamases in the
simulation.
The model is sufficiently generalised so that it
is not specific to a particular drug/target combina-
tion. The key parameters that are required are k
2
, the
rate of inactivation of the target molecule, and K
d
,
the dissociation constant. The ratio of these values
(k
2
/K
d
), or the second order rate constant, is a conve-
nient measure of the drug’s efficacy at inhibiting the
target molecule’s function. The proportion of target
molecule that is inactivated per second, k
a
(the appar-
ent first order rate constant), at a given drug concen-
tration is calculated as a function of these parameters
(equation 2):
k
a
=
k
2
[Ab]
K
d
+ [Ab]
(2)
2.1 Paramaters for Test Simulations
The speed and efficiency of activation of the pro-drug
is an important factor for determining the efficacy of
a pro-drug delivery system. A number of simulations
were carried out to examine the effects of several dif-
ferent parameters on the activation of the pro-drug
and inhibition of bacterial growth by the activated
product. The same cellular parameters for represent-
ing β-lactamase-producing MRSA bacteria that were
used in previously published investigations were ap-
plied here (Table 1) (Murphy et al., 2008; Murphy
et al., 2009). A hypothetical penicillin-based pro-
drug was simulated, i.e. the kinetic parameter val-
ues (k
cat
/K
M
) for penicillin G were used to define
the interaction between β-lactamase and the pro-drug
molecule. The kinetic values for penicillin G were
chosen because this represents a situation where the
β-lactamase enzymes have a high catalytic efficiency
versus the sample pro-drug. This represents an opti-
mal situation in order to assess the potential of this
approach.
The active drug compound that arises from cleav-
age of the simulated penicillin-based pro-drug has ki-
netic parameters (k
2
/K
d
) which determine the rate of
binding to the bacterial cell (Table 1).
Table 1: Inputted parameter values for simulations of
pro-drug interactions with β-lactamase-producing S. au-
reus bacteria in Micro-Gen model. b.u. =biomass units;
loop=program loop (∼2s in real time).
Parameters (units) Input Value
Environment:
Patch size (b.u.) 20000
Patch nutrient level (b.u.) 80000
Diffusion co-efficient 0.1
Bacterial agents:
Generation time (min) 29
Threshold for division (b.u.) 10000
Nutrient intake (b.u. loop
−1
) 10.0
Survival cost (b.u. loop
−1
) 0.2
Stationary phase rel. metabolic rate 0.2
Lag phase length (min) 63
β-lactamase:
Production rate (µM s
−1
): 3.28x 10
−7
Production cost (b.u.) 0.1
Molecular weight (Da) 30000
Half-life (s) 53640
k
cat
(s
−1
): 171.0
K
M
(µM): 51.1
Pro-drug:
Half-life (s) 2520
k
2
(s
−1
): 0.185
K
d
(µM): 1540
3 RESULTS AND DISCUSSION
3.1 Introduction
In order to test the ability of the model to reproduce
real world behaviour of pro-drug compounds a cou-
ple of case studies were carried out previously. These
case studies involved taking kinetic parameters for
two pro-drug compounds from the literature, called
NB2001 and NB2030, and running simulations in or-
der to test the output of the model (Li et al., 2002;
Stone et al., 2004). A detailed description of these
tests is included in a previous publication along with
analysis and discussion of the results (Murphy et al.,
2010). A comparison between the predicted MICs
for NB2001 and NB2030 and the experimentally-
determined values are included in figure 3. The pre-
dicted values matched closely the experimental re-
sults, which indicated a sound theoretical basis for the
model.
However, the power of any modelling approach
does not exist in making predictions, but rather in pro-
viding a basis for a thorough investigation of the dy-
A COMPUTATIONAL MODELLING APPROACH TO EXPLORE THE ANTI-MICROBIAL PRO-DRUG DELIVERY
SYSTEM
303
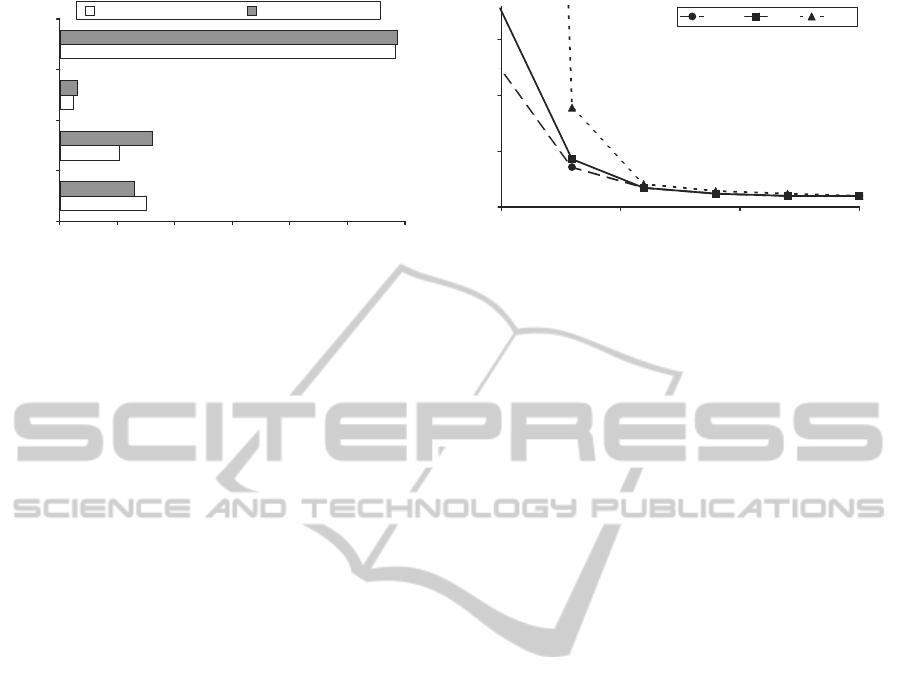
-4 -3 -2 -1 0 1 2
NB2001
NB2030
Triclosan
Pen
Anti-Microbial Drug
Log MIC ( g/ml)
Model Predicted MIC Experimental MIC
Figure 3: Predicted Minimum Inhibitory Concentrations
(MICs) for two pro-drug candidates, NB2001 and NB2030,
described in literature. Predicted MICs for Penicillin G (a
β-lactam antibiotic) and Triclosan (an inhibitor of the bac-
terial fatty acid synthesis cycle, and the active component
of NB2001 and NB2030 (Slater-Radosti et al., 2001)) are
also included for reference.
namics of the system. With that in mind, the focus
of this article is to extend our research to encompass
a more theoretical exploration of the pro-drug system
in order to identify the factors that influence the out-
put. By developing a more holistic understanding of
the pro-drug system, a more rational approach to de-
signing pro-drug candidates can be developed.
3.2 Effect of Kinetic Parameters on
Drug Efficacy
Figure 4 shows the results of tests investigating the
effect of the kinetic parameters on the MIC of the
penicillin-based pro-drug. A lower MIC means a
lower concentration of pro-drug is required in order
to inhibit the bacterial growth. Three hypothetical
variations of the pro-drug were investigated, which
differed by the rate of binding of the activated anti-
microbial agent to its target in the bacterial cell (k
2
/K
d
= 62.5, 250, and 1000 M
−1
s
−1
). The catalytic effi-
ciency of the β-lactamase enzyme (k
cat
/K
M
) at cleav-
ing and activating the pro-drug was assessed by vary-
ing over a range of 10
5
- 10
8
for each pro-drug variant.
As would be expected, higher values for the catalytic
efficiency result in a lower MIC for the pro-drug.
Figure 4 shows that for these pro-drugs the pre-
dicted MIC decreases with increasing catalytic effi-
ciency of the β-lactamase enzyme. The pattern is
the reverse of the trend seen in traditional β-lactam
antibiotics (Murphy et al., 2008). This is one of
the reasons why there is interest in the β-lactamase-
dependent pro-drug delivery system. It would be ex-
pected that administration of β-lactamase-dependent
pro-drugs could lead to evolutionary selective pres-
sure opposed to that exerted by β-lactam antibiotics.
0
5
10
15
1.0E+05 1.0E+06 1.0E+07 1.0E+08
k
cat
/K
M
of -lactamase (M
-1
s
-1
)
Relative MIC of Pro-Drug
1000 250 62.5
Figure 4: Effect of catalytic efficiency (k
cat
/K
M
) of β-
lactamase enzyme on minimum inhibitory concentration
(MIC) of pro-drug. Three different pro-drugs are graphed
which differ by the rate of binding of their activated anti-
microbial agent to the bacterial cell (k
2
/K
d
of activated anti-
microbial agent: 62.5, 250 and 1000 M
−1
s
−1
).
The dynamics between the negativeselective pres-
sure from pro-drugs and positive selective pressure
from β-lactam antibiotics would be an important fac-
tor to consider when assessing the possible evolution
of drug resistance in bacteria in response to these
two different therapeutic strategies. However, the
complex interplay of biophysical, pharmacokinetic,
pharmacological and epidemiological factors which
would contribute to this are beyond the scope of this
study. Nevertheless, this modelling approach is useful
for developing theories about how molecular param-
eters may contribute to the observed dynamics of the
system.
3.3 Effect of β-lactamase Production
Rate on Pro-drug Activation
It is clear that the β-lactamase production rate of the
bacterial cells is an important parameter to be consid-
ered when investigating the β-lactamase-dependent
pro-drug delivery system. The production rate can
vary considerably between different bacterial strains,
and this must be factored in when assessing the
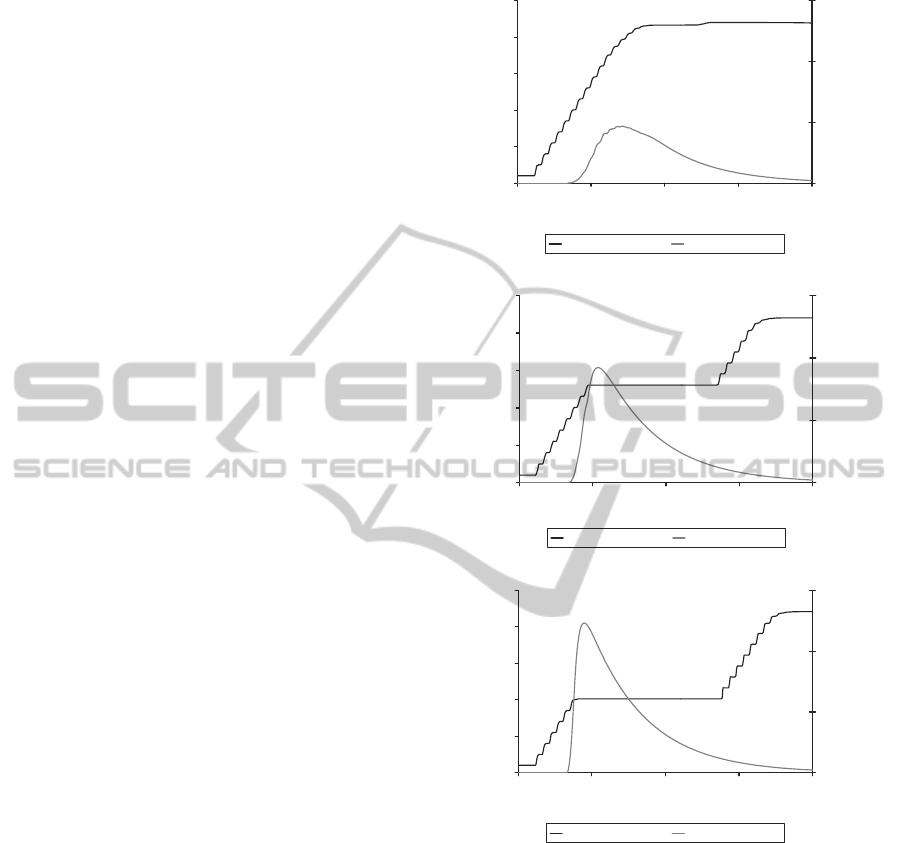
usefulness of this drug delivery system. Figure 5
shows the growth dynamics of a bacterial popula-
tion when exposed to a penicillin-based pro-drug
(1.8µg/ml), with the β-lactamase production rate var-
ied between 10
−7
-10
−5
µM s
−1
agent
−1
For refer-
ence, the β-lactamase production rate for naturally oc-
curring Type A MRSA under these simulation condi-
tions was estimated to be 3.28x 10
−7
µM s
−1
agent
−1
(Murphy et al., 2009).
The results confirm the important role that the
β-lactamase production rate has on the efficacy of
the β-lactamase dependent pro-drug delivery system.
For these simulations, it is assumed that there is no
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
304
contamination or spontaneous activation of the ac-
tive anti-microbial agent apart from activation by β-
lactamase. In real life, there may be some contami-
nation with active compound that would lead to pos-
itive results even when treating β-lactamase-negative
strains of bacteria. However, this ambiguity can lead
to problems when assessing the true effectiveness of
the approach.
The active drug concentration threshold required
for inhibition of growth is approximately 0.8µg/ml.
This threshold is determined by the minimum in-
hibitory concentration of the activated antimicrobial
agent. It is noteworthy however, that increasing the β-
lactamase production rate from 10
−6
to 10
−5
µM s
−1
agent
−1
(fig. 5B -C) does not result in a correspond-
ing increase in the length of time bacterial growth is
inhibited for. The inhibition time seems to be limited
by the half-life of the drug in this case (see fig. 6).
3.4 Effect of Half-life on Pro-drug
Activation
One of the most important parameters that limit the
efficacy of both traditional antibiotics and novel drug
candidates, such as pro-drugs, is the half-life of the
molecule. However, the impact of this parameter can
vary substantially depending on the type of antibi-
otic used (Murphy et al., 2009). The half-life of a
molecule can vary dramatically depending on local
environmental conditions, such as pH or temperature
variations. It is important to determine its influence in
order to attempt to predict treatment success.
For this reason, computational analyses were car-
ried out to predict the impact of this parameter on the
pro-drug delivery system (fig. 6). The half-life of the
simulated pro-drug was varied between 16 minutes
and 2.8 hours, and the growth curve of the bacterial
population plotted along with the concentration curve
of activated anti-microbial agent.
When the half-life is ≤16 minutes, the concentra-
tion of activated anti-microbial agent never exceeds
the threshold required for growth inhibition, 0.8µg/m
(fig. 6A). Therefore, the bacterial population follows
the standard growthcurve, eventuallyentering the sta-
tionary phase due to nutrient limitations. However,
when the half-life of the pro-drug is increased to 1.4
hours or greater, then the required concentration of
active drug compound is reached and inhibition of
growth occurred.
The half-life of a pro-drug is therefore a very
important parameter when determining its efficacy
against β-lactamase-producing S. aureus bacteria.
This system is particulary sensitive to the half-life pa-
rameter because of the time delay between admin-
A.
1.0E+01
1.0E+02
1.0E+03
1.0E+04
1.0E+05
1.0E+06
0 5 10 15 20
Time (hours)
No. of Bacteria
0
0.5
1
1.5
Active Ab Conc.
(
g/ml)
Bac (Lac=E-07) Ab (Lac=E-07)
B.
1.0E+01
1.0E+02
1.0E+03
1.0E+04
1.0E+05
1.0E+06
0 5 10 15 20
Time (hours)
No. of Bacteria
0
0.5
1
1.5
Active Ab Conc.
(
g/ml)
Bac (Lac=E-06) Ab (Lac=E-06)
C.
1.0E+01
1.0E+02
1.0E+03
1.0E+04
1.0E+05
1.0E+06
0 5 10 15 20
Time (hours)
No. of Bacteria
0
0.5
1
1.5
Active Ab Conc.
(
g/ml)
Bac (Lac=E-05) Ab (Lac=E-05)
Figure 5: Effect of varying the β-lactamase production
rate on the inhibition of β-lactamase-producing S. aureus
bacterial growth by the penicillin-based pro-drug. The
graphs display the simulated log bacterial growth curve
along with the concentration of activated drug molecules
(µg/ml). 1.8 µg/ml of pro-drug added at time = 3.3 hours.
β-lactamase production rates: A. 10
−7
µMs
−1
agent
−1
; B.
10
−6
µM s
−1
agent
−1
; C. 10
−5
µMs
−1
agent
−1
.
istration of the pro-drug and activation of sufficient
quantities of active agent to inhibit growth.
A COMPUTATIONAL MODELLING APPROACH TO EXPLORE THE ANTI-MICROBIAL PRO-DRUG DELIVERY
SYSTEM
305

A.
1.0E+01
1.0E+02
1.0E+03
1.0E+04
1.0E+05
1.0E+06
0 10 20 30
Time (hours)
No. of Bacteria
0
0.5
1
1.5
Active Ab Conc.
(
g/ml)
Bac (HL = 16 min) Ab (HL = 16 min)
B.
1.0E+01
1.0E+02
1.0E+03
1.0E+04
1.0E+05
1.0E+06
0 10 20 30 40
Time (hours)
No. of Bacteria
0
0.5
1
1.5
Active Ab Conc.
(
g/ml)
Bac (HL = 1.4 h) Ab (HL = 1.4 h)
C.
1.0E+01
1.0E+02
1.0E+03
1.0E+04
1.0E+05
1.0E+06
0 10 20 30 40
Time (hours)
No. of Bacteria
0
0.5
1
1.5
Active Ab Conc.
(
g/ml)
Bac (HL = 2.8 h) Ab (HL = 2.8 h)
Figure 6: Effect of varying the half-life parameter for the
penicillin-based pro-drug on the inhibition of β-lactamase-
producing S. aureus bacterial growth. The graphs display
the simulated log bacterial growth curve along with the con-
centration of activated drug molecules (µg/ml). 1.8 µg/ml
pro-drug added at time = 3.3 hours. A. Half-life=16min;
B. Half-life= 1.4 h; C. Half-life=2.8 h.
3.5 Effect of Diffusion Rate on
Pro-Drug Activation
The impact of diffusion on the activity and dynamics
of the pro-drug delivery system was also investigated.
This is an important parameter to assess because the
pro-drug delivery system depends on the targeted re-
lease of active anti-microbialagents in the close vicin-
ity of bacterial cells. The implementation of an algo-
rithm based on Fick’s First Law of diffusion allows
some insights to be obtained on the role of diffusion
dynamics in the system. The agent-based approach
allows us to explicitly take into account spatial hetero-
geneity in the environmental conditions (e.g. between
the inside and outside of the colony) which is impor-
tant when considering features such as diffusion.
Figure 7 shows the impact of varying the rate of
diffusion in the environmenton the activation and effi-
cacy of a penicillin-based pro-drug. The rate of diffu-
sion was varied by modifying the user-defined diffu-
sion coefficient (D) for Fick’s First Law of diffusion.
This is a system-level parameter that alters the rates
of diffusion of all the molecules (pro-drug, active
drug, β-lactamase and nutrients) in the environment.
Higher values correspond to a more fluid/dynamic en-
vironment whereas lower values result in a more vis-
cous/inert simulated environment. The results from
these tests indicate that the rate of diffusion has an im-
portant influence on the availability of activated pro-
drug in the vicinity of the bacterial cells as measured
by the height of the peak in the concentration of acti-
vated anti-microbial agent (fig. 7).
When the diffusion rate was lower (fig. 7A), the
concentration of activated anti-microbial agents in the
local vicinity of the bacterial cells increased more
rapidly. This may be explained by the fact that with
lower diffusion rates, the activated drug molecules
would not be dispersed as quickly from the vicinity
of the bacterial cells by natural diffusion processes
(fig. 8A).
The pro-drug delivery system results in higher
concentrations of activated drug molecules in the di-
rect vicinity of the bacterial cells. The system is there-
fore sensitive to any forces, such as diffusion or flow
forces, that may result in dispersal of the activated
compounds. It is important, therefore, to take this
into account when designing pro-drugs and try to take
measures to minimize this such as, for example, de-
signing molecules that have a greater binding affinity
or are electrostatically attracted to the bacterial cells.
This problem is not so evident with traditional an-
tibiotic approaches because they usually involve the
administration of relatively high doses of active anti-
microbial agent that are not specifically targeted to the
local vicinity of the bacterial cells. In fact for some
types of β-lactam antibiotic, such as penicillin G, in-
creasing the diffusion rate was predicted to increase
antibiotic efficacy (Murphy et al., 2009). This could
be due to the fact that higher rates of diffusion results
in dispersal of β-lactamase-inactivated penicillin G in
the vicinity of the bacterial cells and replacement by
active penicillin G from elsewhere in the environment
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
306
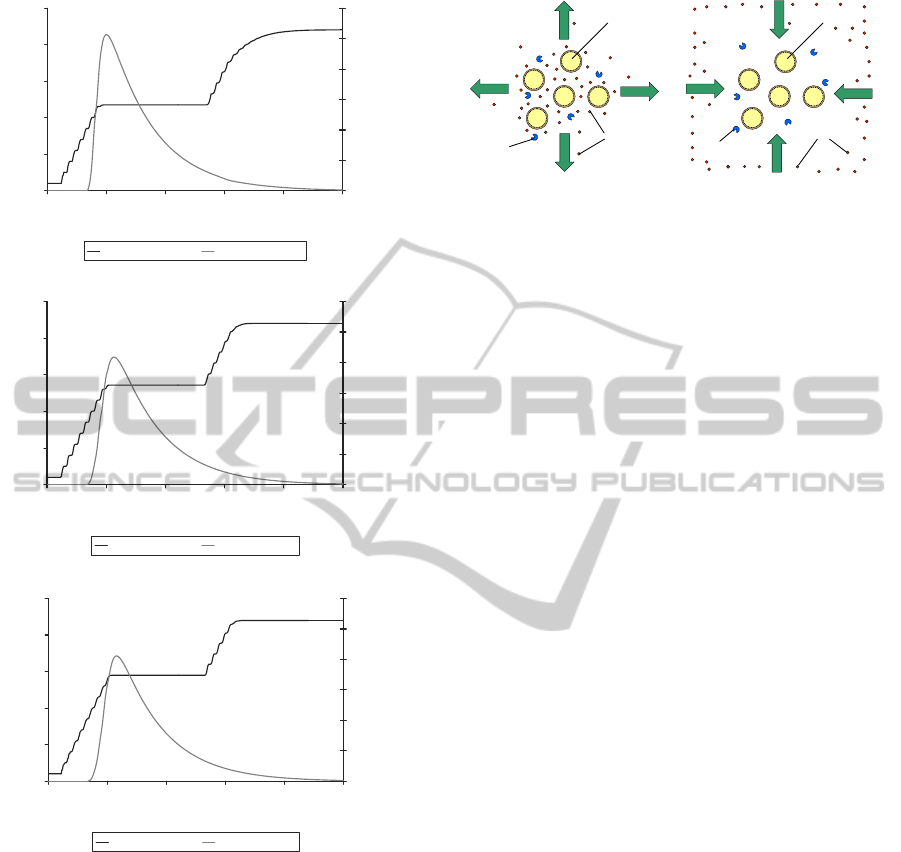
A.
1.0E+01
1.0E+02
1.0E+03
1.0E+04
1.0E+05
1.0E+06
0 5 10 15 20 25
Time (hours)
No. of Bacteria
0
0.2
0.4
0.6
0.8
1
1.2
Active Ab Conc.
(
g/ml)
Bac (Dif=0.01) Ab (Dif=0.01)
B.
1.0E+01
1.0E+02
1.0E+03
1.0E+04
1.0E+05
1.0E+06
0 5 10 15 20 25
Time (hours)
No. of Bacteria
0
0.2
0.4
0.6
0.8
1
1.2
Active Ab Conc.
(
g/ml)
Bac (Dif=0.1) Ab (Dif=0.1)
C.
1.0E+01
1.0E+02
1.0E+03
1.0E+04
1.0E+05
1.0E+06
0 5 10 15 20 25
Time (hours)
No. of Bacteria
0
0.2
0.4
0.6
0.8
1
1.2
Active Ab Conc.
(
g/ml)
Bac (Dif=0.2) Ab (Dif=0.2)
Figure 7: Effect of varying the user-defined diffusion pa-
rameter (D) for Fick’s First Law of diffusion on the inhibi-
tion of β-lactamase-producing S. aureus bacterial growth by
the penicillin-based pro-drug. The graphs display the sim-
ulated log bacterial growth curve along with the concentra-
tion of activated drug molecules (µg/ml). 1.8 µg/ml of pro-
drug added at time = 3.3 hours. A. D = 0.001; B. D = 0.01;
C. D =0.1.
- the reversesituation to the pro-drugsystem (fig. 8B).
4 CONCLUSIONS
This paper documents tests to analyse the system dy-
namics of the β-lactamase-dependent therapeutic ac-
A. B.
S. aureus cell
Active Ab
molecules
Direction of diffusion
of active Ab
-lactamase
enzyme
S. aureus cell
Active Ab
molecules
Direction of diffusion
of active Ab.
-lactamase
enzyme
Figure 8: Comparison of the local diffusion gradients of
active antimicrobial agents in the local environment of β-
lactamase producing S. aureus cells for pro-drug (A) and
traditional β-lactam antibiotics (B). A. When administered
in pro-drug form the active antimicrobial agent concentra-
tion is highest in the vicinity of the bacterial cells due to β-
lactamase-mediated activation. B. For traditional β-lactam
antibiotics the concentration of active agent is depleted in
the vicinity of the bacterial cells due to inactivation by the
β-lactamases and uptake by the cells.
tivation pro-drug delivery system, a novel approach
for achieving β-lactamase-mediated selective release
of antimicrobial agents. It is thought that this strat-
egy might be a promising approach for treating β-
lactamase over-expressing strains of bacteria that are
resistant to traditional β-lactam antibiotics. The ini-
tial results are promising and illustrate the power of
the computational approach for exploring the mech-
anisms of action of novel drug compounds. In con-
junction with laboratory testing, great insights can
be made into the complex interplay of the different
components in the pro-drug delivery system using an
agent-based modelling approach. Initial work has al-
ready been carried out on data from real-life pro-drug
candidates in order to compare the model output to
experimental results (Murphy et al., 2010). How-
ever, the power of the model exists in being able to
explore hypothetical scenarios and compounds in or-
der to gain an integrated understanding of the unique
dynamics of the pro-drug delivery system. By using
different modelling approaches to inform decisions in
the rational drug design process it is possible to opti-
mize the effectiveness of this technique so as to offer
a viable alternative treatment strategy for microbial
infectious diseases.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the contribu-
tion of Mathieu Joubert, Grainne Kerr, Chris Pender,
Marian Duggan and Ronan Winters for the develop-
ment of the original BAIT software tool. High per-
formance computing resources were provided by the
Centre for Scientific Computing and Complex Sys-
A COMPUTATIONAL MODELLING APPROACH TO EXPLORE THE ANTI-MICROBIAL PRO-DRUG DELIVERY
SYSTEM
307

tems Modelling (SCI-SYM) in Dublin City Univer-
sity, and the Irish Centre for High End Computing
(ICHEC).
REFERENCES
Abraham, E. P. and Chain, E. (1940). An enzyme from
bacteria able to destroy penicillin. Nature, 146:837.
Bush, K., Macielag, M., and Weidner-Wells, M. (2004).
Taking inventory: antibacterial agents currently at or
beyond phase 1. Current opinion in microbiology,
7(5):466–476.
Fisher, J. F., Meroueh, S. O., and Mobashery, S. (2005).
Bacterial resistance to beta-lactam antibiotics: com-
pelling opportunism, compelling opportunity. Chemi-
cal reviews, 105(2):395–424.
Ginovart, M., Lopez, D., and Valls, J. (2002). Indisim, an
individual-based discrete simulation model to study
bacterial cultures. Journal of theoretical biology,
214(2):305–319.
Jennings, N. R., Sycara, K., and Wooldridge, M. J. (1998).
A roadmap of agent research and development. Au-
tonomous Agents and Multi-Agent Systems, 1(1):7–
38.
Levy, S. B. and Marshall, B. (2004). Antibacterial resis-
tance worldwide: causes, challenges and responses.
Nature medicine, 10(12 Suppl):S122–9.
Li, Q., Lee, J. Y., Castillo, R., Hixon, M. S., Pujol, C.,
Doppalapudi, V. R., Shepard, H. M., Wahl, G. M.,
Lobl, T. J., and Chan, M. F. (2002). Nb2001, a novel
antibacterial agent with broad-spectrum activity and
enhanced potency against beta-lactamase-producing
strains. Antimicrobial Agents and Chemotherapy,
46(5):1262–1268.
Murphy, J. T., Walshe, R., and Devocelle, M. (2007).
Agent-based model of methicillin-resistant staphylo-
coccus aureus and antibiotics in batch culture. In
Proceedings of 21st Annual European Simulation and
Modelling Conference, pages 409–414. Eurosis-ETI.
Murphy, J. T., Walshe, R., and Devocelle, M. (2008). A
computational model of antibiotic-resistance mecha-
nisms in methicillin-resistant staphylococcus aureus
(mrsa). Journal of theoretical biology, 254(2):284–
293.
Murphy, J. T., Walshe, R., and Devocelle, M. (2009). Mod-
elling the population dynamics of antibiotic-resistant
bacteria: An agent-based approach. International
Journal of Modern Physics C, 20(3):435–457.
Murphy, J. T., Walshe, R., and Devocelle, M. (2010). A
theoretical analysis of the pro-drug delivery system
for treating antibiotic-resistant bacteria. IEEE/ACM
Transactions on Computational Biology and Bioinfor-
matics, 99.
Neu, H. C. (1992). The crisis in antibiotic resistance. Sci-
ence, 257(5073):1064–1073.
Rautio, J., Kumpulainen, H., Heimbach, T., Oliyai, R., Oh,
D., Jarvinen, T., and Savolainen, J. (2008). Prodrugs:
design and clinical applications. Nature reviews.Drug
discovery, 7(3):255–270.
Slater-Radosti, C., Aller, G. V., Greenwood, R., Nicholas,
R., Keller, P. M., Jr, W. E. D., Fan, F., Payne, D. J., and
Jaworski, D. D. (2001). Biochemical and genetic char-
acterization of the action of triclosan on staphylococ-
cus aureus. The Journal of antimicrobial chemother-
apy, 48(1):1–6.
Smyth, T. P., O’Donnell, M. E., O’Connor, M. J.,
and StLedger, J. O. (2000). beta-lactamase-
dependent prodrugsrecent developments. Tetrahe-
dron, 56(31):5699–5707.
Stone, G. W., Zhang, Q., Castillo, R., Doppalapudi, V. R.,
Bueno, A. R., Lee, J. Y., Li, Q., Sergeeva, M., Kham-
batta, G., and Georgopapadakou, N. H. (2004). Mech-
anism of action of nb2001 and nb2030, novel antibac-
terial agents activated by beta-lactamases. Antimicro-
bial Agents and Chemotherapy, 48(2):477–483.
Zygmunt, D. J., Stratton, C. W., and Kernodle, D. S. (1992).
Characterization of four beta-lactamases produced by
staphylococcus aureus. Antimicrobial Agents and
Chemotherapy, 36(2):440–445.
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
308
