
SILICON-BASED GOLD TRANSDUCERS FOR DNA
BIOSENSORS
Łukasz Górski, Robert Ziółkowski, Elżbieta Malinowska
Warsaw University of Technology, Faculty of Chemistry, Department of Microbioanalytics, Noakowskiego 3, 00-664
Warsaw, Poland
Piotr Prokaryn, Michał Zaborowski
Institute of Electron Technology, Al. Lotników 32/46, 02-668 Warsaw, Poland
Keywords: Voltammetry, Vacuum deposited gold, DNA biosensors, Monolayers.
Abstract: Silicon-based chips with vacuum deposited gold electrode were tested as transducers for the development of
DNA-modified biosensors. It was found that these structures are superior over commercially available
transducers, mainly due to perfectly smooth surface of gold working electrode. This was confirmed with
microscopic and electrochemical experiments. Obtained transducers were modified with oligonucleotide
self-assembled monolayer. These sensors were shown to detect chosen DNA sequence with the employment
of methylene blue as a redox marker. The same sensors were used to determine UO
2
2+
cation, however these
efforts were unsuccessful.
1 INTRODUCTION
The still-growing number of data which can be used
in healthcare, powers the continuing research over
the development of new, accurate and cheap
methods for the analysis of biological compounds
(Luong, Male, Glennon, 2008; Andreescu, Sadik,
2004). The concerns over analysis costs are also
very important, as the majority of the clinical
analysis are performed in great numbers every day.
Various analytical techniques have been used in
clinical analysis, each of them having its strengths
and drawbacks. The ease of miniaturization,
reduction of reagent consumption as well as high
sensitivity, reproducibility and accurate analysis are
the reasons for high interest in electrochemistry
(Wang, 2000). However, to take advantage of all
these benefits, the appropriately constructed and
mass produced transducers, with well defined
surface, allowing for an easy and reproducible
creation of the (bio)recognition layer, are required.
Apart from the proper transducer, receptor layer
is extremely important for sensor performance. Short
oligonucleotides, known as aptamers, have recently
gained attention as promising receptors for the
construction of novel biosensors. The nucleotides
sequence of the DNA strand may be the indicator of
several health-important problems, blood
contamination with pathogenic bacteria or
genetically modified food, thus DNA sequence is
one of the most interesting target for aptasensors
(Hianik, Wang, 2009). However, the detection of
various small biomolecules, as well as inorganic
ions can be performed by sensors modified with
oligonucleotides.
Another important component, typically present
in electrochemical sensors, is a redox marker. In
DNA sensors, markers are usually responsible for
detection of hybridization or interaction of an
aptamer with analyte. There are three possible
modes for binding of a marker with DNA, including
electrostatic, groove and intercalative binding
(Erdem, Ozsoz, 2002). Among compounds
employed as electrochemical DNA markers,
methylene blue is probably most popular, however
the design and application of new redox markers is
still an important research topic.
In this work, a holistic approach to the design
and fabrication of various aptasensors is presented.
The usefulness of the silicon-based transducers with
vacuum deposited gold for the construction of such
sensors is evaluated. Various geometries of
146
Górski Ł., Ziółkowski R., Malinowska E., Prokaryn P. and Zaborowski M..
SILICON-BASED GOLD TRANSDUCERS FOR DNA BIOSENSORS.
DOI: 10.5220/0003133901460149
In Proceedings of the International Conference on Biomedical Electronics and Devices (BIODEVICES-2011), pages 146-149
ISBN: 978-989-8425-37-9
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

transducers were developed and tested, including
back-side contact chips and dipstick three-electrode
structures. The performance of these transducers was
compared with typical gold disc electrodes and
commercially available three-electrode structures.
The modification of silicon-based chips with
oligonucleotide layer, based on self-assembling
process, was also performed. Obtained sensors were
used for detection of chosen DNA sequence.
Moreover, the efforts to determine UO
2
2+
cation
concentration using similar biosensors, will be
described.
2 EXPERIMENTAL
2.1 Apparatus
Electrochemical measurements were conducted with
a CHI 660A electrochemical workstation (CH
Instruments, USA). Voltammetric experiments were
carried out with a three-electrode system. If applied,
external auxiliary electrode was a gold wire, while
Ag/AgCl/1.0 M KCl was used as an external
reference electrode (Mineral, Poland). The sample
solutions were deoxygenated with argon for
approximately 15 minutes prior to data acquisition
and were blanketed under an argon atmosphere
during the entire experimental period. The square
wave voltammetry (SWV) was conducted at a pulse
amplitude of 25 mV, step of 1 mV and frequency of
25 Hz. The potentials of chronoamperometry (CA)
assay were changed from the initial E = -200 mV to
E = 900 mV and it was kept constant for 0.5 sec.
The surface pictures were taken by the TM-1000
electron microscope (HITACHI, Japan) and the
stereomicroscope SZX10 (Olympus, Japan) coupled
with CCD color camera ColorView (Olympus,
Japan).
As working electrodes, silicon-based gold
transducers with back-side contact (BSC)
(Ziolkowski et al., 2010) and vacuum deposited
gold, as well as 3-electrode structures (TES) based
on silicone wafer, with vacuum deposited gold
working electrode (Institute of Electron Technology,
Poland), were used (Figure 1). For fabrication of
these transducers, a planar CMOS-compatible
process was used. For comparison, typical gold disc
electrode (Mineral, Poland) (GDE) and
commercially available transducers, based on
ceramic support, with screen printed gold (SPG)
working electrode (BVT Technologies, Czech
Republic), were also tested.
Figure 1: Silicon-based transducers used in this work: A –
three-electrode structure (TES) (dimensions 7x26 mm); B
– back-side contact structures (dimensions 5x5 mm).
2.2 Reagents
Analytical-grade K
4
Fe(CN)
6
, K
3
Fe(CN)
6
, KCl,
K
2
HPO
4
, KH
2
PO
4
, NaCl, NaOH, HCl, Tris-HCl,
methylene blue, UO
2
(NO
3
)
2
and ascorbic acid were
purchased from Aldrich Chemicals. Absolute
ethanol was purchased from POCh, Poland. All
reagents were used without further purification. All
solutions were prepared using Milli-Q water. Milli-
Q water and all buffers were sterilized using an
autoclave. The 20-mer deoxyoligonucleotides were
purchased from Genomed Sp. z o. o., Poland. The
base sequences were as follows:
- thiolated DNA probe: 5'-SH-(CH2)6-
TCCAACACTCCGAGACGGGG-3'
- complementary DNA target 5'-
CCCCGTCTCGGAGTGTTGGA-3'.
All oligonucleotide stock solutions were
prepared with 10mM Tris–HCl, (pH 7.5) and stored
in a −20 ºC freezer before use.
2.3 Solutions
The following solutions were prepared: piranha
solution (H
2
O
2
:H
2
SO
4
; 3:1); 0.01 M solution of
K
4
Fe(CN)
6
and K
3
Fe(CN)
6
in 0.1M KCl;
immobilization buffer solution containing 1 M
KH
2
PO
4
(pH 4.5); hybridization buffer solution
containing 10 mM Tris–HCl and 1 M NaCl (pH 7.0).
Buffer solution for electrochemical measurements
was composed of 0.05M K
2
HPO
4
/KH
2
PO
4
and 0.3
M NaCl (pH 7.0). The 10 μM methylene blue stock
solution was prepared in the electrochemical buffer.
The pH was adjusted with either NaOH or HCl
solution.
A
B
SILICON-BASED GOLD TRANSDUCERS FOR DNA BIOSENSORS
147
2.4 Methods
The pictures of the surfaces were taken according to
the microscope producers’ instructions (HITACHI,
Olympus, Japan).
The real surface area was calculated using the
Cottrell equation with the data from
chronoamperometric experiments.
To prepare DNA-modified sensors, transducers
were cleaned with piranha solution and
electrochemically prepared by cycling the potential
scan between –0.6 and 1.8V at the scan rate of 0.1V
s
-1
in electrochemical buffer. Subsequently, drop of
4 μM thiolated ssDNA probe in immobilization
buffer was placed on the electrode surface and the
whole chip was placed in the Petri dish lined with a
blotting paper soaked with immobilization buffer.
The immobilization was carried on for 90 min at
room temperature. This recognition interface was
then electrochemically examined with methylene
blue solution (Kelley et al., 1997). After analysis,
the methylene blue was washed away and the
modified gold electrode was exposed to sample
solution. After that time, chips were rinsed with
phosphate buffer and again the electrochemical
examination in methylene blue solution was
conducted.
3 RESULTS AND DISCUSSION
3.1 Transducers
To evaluate the surface structure of prepared silicon-
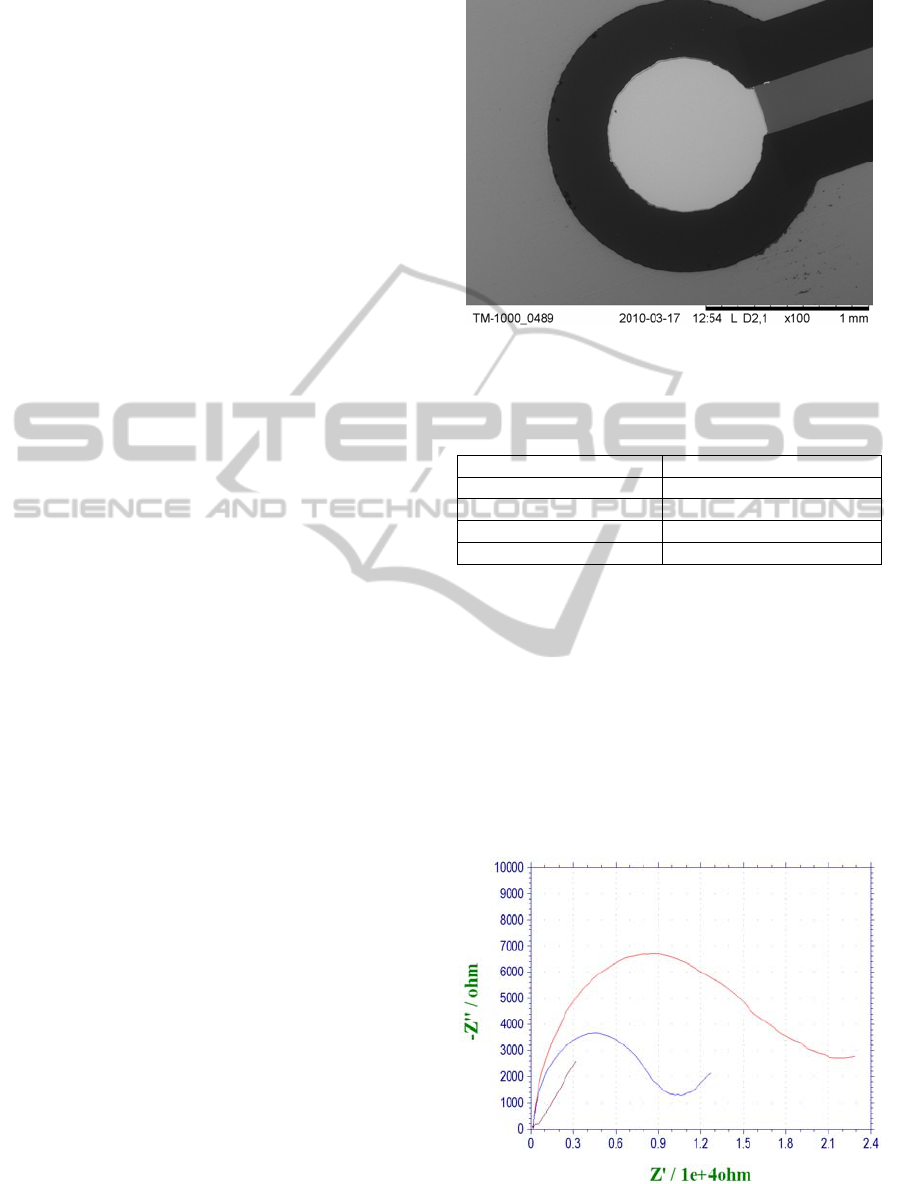
based electrodes, optical and electron microscopy
was employed. The pictures from electron
microscope showed the heterogeneity of the
electrode in the case of SPG sensors and the
smoothness and integrity in the case of BSC and
TES structures (Figure 2). For the classical gold disc
electrode, only optical pictures were taken (data not
shown), showing its significant unevenness, clearly
visible even at relatively low (10x) magnification.
As a consequence, the electrochemical area,
measured using chronoamperometry in
K
4
Fe(CN)
6
/K
3
Fe(CN)
6
solution, was quite similar to
geometric area for silicon-based transducers. Table 1
shows the roughness factor for each type of
electrode used in the present study. It is evident that
BSC and TES transducers have better defined
electrode surface, as compared to commercially
available GDE and SPG.
Figure 2: Electron microscope image of a working
electrode of three-electrode structure (TES).
Table 1: Roughness factor for gold electrodes of
transducers tested in this work.
Transducer Roughness factor
GDE 3.45
SPG 2.61
BSC 1.75
TES 1.02
3.2 DNA Detection
To devise DNA biosensor constructed with
fabricated BSC and TES transducers, the
immobilization of ssDNA and subsequent
hybridization with complementary strand was
observed using impedance spectroscopy. The gold
electrode was prepared according to procedure
described in experimental section. Figure 3 shows
impedance spectra of TES transducer with working
electrode modified with ssDNA, as well as for the
Figure 3: Impedance spectra of TES transducer with: A –
bare gold electrode; B – DNA receptor layer; C – after
hybridization.
A
B
C
BIODEVICES 2011 - International Conference on Biomedical Electronics and Devices
148

same sensors after the hybridization. Based on these
results, detection of hybridization event is evident.
Similar experiment could not be carried out for
BSC sensors, most probably due to high impedance
through the doped silicon structure. Accordingly,
hybridization was detected using square wave
voltammetry (SWV), using methylene blue as a
redox marker. Again, obtained data confirm the
immobilization of ssDNA on the gold surface, as
indicated by redox potential shift (ΔE = 0.013V).
Further shift in redox potential (ΔE = 0.028V)
confirms that hybridization of receptor DNA layer
with sample ssDNA takes place.
3.3 UO
2
2+
Detection
It was reported recently that uranyl cation can cause
DNA damage, especially in the presence of ascorbic
acid (AA) (Yazzie et al., 2003). Many people can be
potentially exposed to UO
2
2+
through uranium
mining, processing, the resulting mine tailings, and
the use of depleted uranium in the military. Thus,
determination of uranyl cation is very important
from the clinical point of view. Based on the
reported cleavage effect of UO
2
2+
/AA on DNA,
efforts were undertaken to devise uranyl sensor
taking advantage of the degradation of DNA self-
assembled monolayer, deposited on the BSC
transducers. Surprisingly, UO
2
2+
had no effect on the
monolayer, even in the presence of ascorbic acid, as
observed using impedance spectroscopy and SWV.
Currently, work is in progress in our laboratory to
elucidate this unsuspected behavior.
4 CONCLUSIONS
Silicon-based transducers with vacuum deposited
gold were found to be useful for the construction of
DNA sensors, mainly due to perfectly smooth
surface of gold working electrode. Produced sensors,
modified with oligonucleotide self-assembled
monolayer, were shown to detect chosen DNA
sequence. Efforts to determine UO
2
2+
cation using
the same sensors were unsuccessful.
ACKNOWLEDGEMENTS
This work was financed by the Polish Ministry of
Science and Higher Education within a framework
of the Operational Programme – Innovative
Economy, Priority I, Action 1.3, Sub-Action 1.3.1,
Project No. POIG.01.03.01-00-014/08-00.
REFERENCES
Luong, J. H. T., Male, K. B., Glennon, J. D., 2008.
Biosensor technology: Technology push versus market
pull. In Biotechnol. Adv.
Andreescu, S., Sadik O. A., 2004. Trends and challenges
in biochemical sensors for clinical and environmental
monitoring. In Pure Appl. Chem.
Wang, J., 2000. Analytical electrochemistry, Wiley-VCH.
New York, 2
nd
edition.
Hianik, T., Wang, J., 2009. Electrochemical Aptasensors -
Recent Achievements and Perspectives. In
Electroanalysis.
Erdem, A., Ozsoz, M., 2002. Electrochemical DNA
Biosensors Based on DNA-Drug Interactions. In
Electroanalysis.
Ziolkowski, R., Gorski, L., Zaborowski, M., Malinowska,
E., 2010. Application of mass fabricated silicon-based
gold transducers for amperometric biosensors. In
Bioelectrochemistry.
Kelley, S.O., Barton, J. K., Jackson, N. M., Hill, M. G.,
1997. Electrochemistry of Methylene Blue Bound to a
DNA-Modified Electrode. In Bioconjugate Chem.
Yazzie, M., Gamble, S. L., Civitello, E. R., Stearns D. M.,
2003. Uranyl Acetate Causes DNA Single Strand
Breaks In Vitro in the Presence of Ascorbate (Vitamin
C). In Chem. Res. Toxicol.
SILICON-BASED GOLD TRANSDUCERS FOR DNA BIOSENSORS
149
