
AUTOMATIC SEGMENTATION OF EMBRYONIC HEART IN
TIME-LAPSE FLUORESCENCE MICROSCOPY IMAGE
SEQUENCES
P. Krämer, F. Boto
1
, D. Wald, F.Bessy, C. Paloc
Vicomtech, Paseo de Mikeletegi 57, 20009 Donostia, San Sebastián, Spain
C. Callol
2
, A. Letamendia, I. Ibarbia, O. Holgado, J. M. Virto
Biobide, Paseo de Mikeletegi 58, 20009 Donostia, San Sebastián, Spain
Keywords: Segmentation, Fluorescent microscopy images, Embryonic heart.
Abstract: Embryos of animal models are becoming widely used to study cardiac development and genetics. However,
the analysis of the embryonic heart is still mostly done manually. This is a very laborious and expensive
task as each embryo has to be inspected visually by a biologist. We therefore propose to automatically
segment the embryonic heart from high-speed fluorescence microscopy image sequences, allowing
morphological and functional quantitative features of cardiac activity to be extracted. Several methods are
presented and compared within a large range of images, varying in quality, acquisition parameters, and
embryos position. Although manual control and visual assessment would still be necessary, the best of our
methods has the potential to drastically reduce biologist workload by automating manual segmentation.
1 INTRODUCTION
Model organisms have become more and more
important for the study of vertebrate development.
Due to its prolific reproduction and the external
development of the transparent embryo, they are
prime models for genetic and developmental studies,
as well as research in toxicology and genomics.
While genetically more distant from humans, the
vertebrate models nevertheless have comparable
organs and tissues, such as heart, kidney, pancreas,
bones, and cartilage.
During the last years tremendous advances in
imaging system have been made allowing the
acquisition of high-resolution images of embryos.
Anyhow, the processing of such images is still a
challenge (Vermot et al., 2008). To date, only little
work has been presented addressing the analysis of
embryo models images (Fink et al., 2009; Luengo-
Oroz et al., 2007; Liebling et al., 2006). For instance
(Liebling et al., 2006) presents a method to acquire,
reconstruct and analyze 3D images of the zebrafish
heart. The reconstruction of the volume is based on a
semi-automatic segmentation procedure and requires
the help of the user. Fink et al., 2009 propose a
method for detection and quantification of heartbeat
parameters in Drosophilia deriving a signal from the
images, avoiding segmentation.
In case of studies of cardiac development, a
segmentation of the heart provides additional
information for its quantification. Therefore, we
present several approaches to automatically extract
its shape and each chamber from image sequence. In
our experiment, transgenic embryos expressing
fluorescent protein in the myocardium were placed
under light microscopy allowing to capture
fluorescent images of the heart at video rate. In
particular, we are interested in segmenting the heart
of zebrafish embryos after two days of post-
fertilization (2 dpf). In early stages of the zebrafish
development the primitive heart begins a simple
linear tube. This structure gradually forms into two
chambers, a ventricle and an atrium. At 2 dpf the
heart tube is already partitioned into atrium and
ventricle as depicted in Figure 1. They are separated
by a constriction which will later form the valve. At
this stage the heart is already beating. More
information on zebrafish heart anatomy can be found
121
Krämer P., Boto F., Wald D., Bessy F., Paloc C., Callol C., Letamendia A., Ibarbia I., Holgado O. and Virto J. (2010).
AUTOMATIC SEGMENTATION OF EMBRYONIC HEART IN TIME-LAPSE FLUORESCENCE MICROSCOPY IMAGE SEQUENCES.
In Proceedings of the Third International Conference on Bio-inspired Systems and Signal Processing, pages 121-126
DOI: 10.5220/0002342501210126
Copyright
c
SciTePress

in (Hu et al., 2000).
The remainder is organized as follows: In section
2 we present several approaches to segment the
zebrafish heart and in section 3 two methods to
identify the chambers. In section 4 we show some
results and compare the segmentation methods
respectively for the heart and its chambers. We give
a conclusion of our work and outline future research
in section 5.
Figure 1: The 2 dpf zebrafish heart already consists of two
chambers: the atrium (A) and ventricle (V).
2 SEGMENTATION OF THE
ZEBRAFISH HEART
In this section we outline different approaches to
segment the shape of the zebrafish heart. For the
methods of subsection 2.3, 2.4, 2.5, we cast the
images to 8-bit grey level images and stretch the
grey level range into [0,255].
2.1 Adaptive Binarization
This method is based on the assumption that the
image of the heart consists of three brightness levels
such as illustrated in Figure 2: one corresponding to
the background and two corresponding to the
fluorescent heart where strong contracted regions
appear brighter due to a higher concentration of
fluorescent cells.
For pre-processing, we smooth image using a
Gaussian filter to remove noise. Then, the region of
the heart with highest brightness is segmented by
first applying a Contrast-Limited Adaptive
Histogram Equalization (CLAHE) (Zuiderveld,
1994) using a uniform transfer function and then the
automatic threshold method from Otsu (Otsu, 1979).
In order to segment the second, less brighter region
of the heart, we exclude the previous segmented
Figure 2: The image of fluorescent heart consists of three
brightness levels: one corresponding to the background
and two to the heart.
region and apply CLAHE and Otsu again. The
final segmentation is obtained by combining both
segmentation results. Postprocessing includes the
filling of holes which can appear inside in the shape.
2.2 Clustering
This method is based on unsupervised classification
in order to distinguish between object and
background pixels. First, each pixel is characterized
by the mean luminance value of the 3×3 mask
centered at the pixel. A unidimensional feature space
results. Then, we use a k-means classifier (k=3) in
order to separate the pixels into three clusters. This
method relies like the previous one on the
assumption than that there are three different
brightness levels. The cluster to which belongs the
pixel at position (0,0) is then defined as the
background and others as the region of the heart.
Similarly than above, we apply hole filling as
postprocessing. For more information on k-means
clustering can be found in (Bishop, 2007).
2.3 Voronoi-based Segmentation
The Voronoi segmentation (Imelinska et al., 0002) is
based on repeatedly dividing an image into regions
using Voronoi diagram and classifying the regions
as either inside or outside the target based on
classification statistics, and then break up the
regions on the boundary between the two
classifications into smaller regions and repeat the
BIOSIGNALS 2010 - International Conference on Bio-inspired Systems and Signal Processing
122

classification and subdivision on the new set of
regions. The classification statistics can be obtained
from an image prior which is a binary image of
preliminary segmentation.
In order to compute the image prior, we apply
first a bilateral filter to smooth the image while
preserving edges. Afterwards, the gradient
magnitude is computed using a recursive Gaussian
filter and Sigmoid filter to map the intensity range
into [0,255]. Then a threshold is applied to the
gradient magnitude to obtain a binary mask. As the
binary mask may contain holes, we apply a
morphological closing operation and fill the holes to
complete the object’s shape. Then the main region of
the heart is isolated from noise in the binary image
by a region growing algorithm to the binary with the
brightest pixel in the image as seed point. Typically,
the brightest pixel in the gray-level image belongs to
the region of the heart. After the Voronoi
segmentation we apply again morphological closing,
hole filling, isolation of the main region, and
morphological erosion to smooth the contours.
2.4 Level Set
The idea of this method is similar to the previous
one. First a pre-segmentation accomplished which is
then refined, but here we use the level set approach
(Li et al., 2005) for refinement. We choose this
method because of its fast performance.
The method starts with a morphological
reconstruction to suppress structures that are lighter
than their surroundings and that are connected to the
image border. Then, edges are detected using the
Canny edge detector. Dilation, hole filling, and
erosion are applied to the contour image. The
biggest region is considered as the region of the
heart while the others are considered as noise. We
complete the form by applying again dilation and
hole filling.
A Gaussian filter is applied to smooth the
original grey level image for noise removal. Then
we apply the level set method (Li et al., 2005) with
contours of the binary mask as initialization. We
chose the edge indicator function 1/(1+g) as
suggested by the author where g is the gradient
magnitude of the Gaussian filtered grey level image.
2.5 Watershed
This approach is different to the previous one as it
does not rely on a pre-segmentation by binarization.
It is based on Watershed segmentation.
First the border structures are supressed by
morphological reconstruction. This is followed by a
strong low-pass filtering (Gaussian filter) in a
morphological reconstruction by erosion using the
inverse of morphological gradient. This attenuates
unwanted portions of the signal while maintaining
the signal intensity as the Watershed method is
known to oversegment the image. Afterwards, a
small threshold is applied to set the background to
zero and the image intensity is adjusted so that such
that 1% of data is saturated at low and high
intensities. We apply to this gradient magnitude the
watershed segmentation. An oversegmented image
may result with typically one region belonging to the
background and several regions belonging to the
heart. The latter ones are joined to form the region of
the heart.
3 IDENTIFICATION OF THE
CHAMBERS
The objective is now to divide the heart into the
chambers based on the results of the methods
presented in the previous section.
3.1 Convexity Defects
The method assumes that there is a constriction
between the two chambers (see Figure 1) causing
two convex points in the contour of the heart’s
shape. Therefore, we compute the convexity defects
of the contour using its convex hull. Generally more
than two convexity defects are found due to
irregularities in the contour caused by the
segmentation as depicted in Figure 3. Moreover, we
assume that the convexity defects denoting the
constriction between the chambers are parallel.
Thus, we choose the four most important convexity
defects, i.e. the four points with the highest distance
from the convex hull, and compute the angle for
each pair as:
.
v
|
||
v
|
(1)
where v1,v2 are respectively the vectors between the
start and end points of the first and second convexity
defects. If the angle is lower than a small threshold,
then the pair of convexity defects is considered as a
possible candidate for the constriction, otherwise it
is rejected. Finally, we choose the pair with the
highest mean distance as the points of the
constriction from the remaining. We compute the
straight line interpolating the points which separates
both chambers. As there is often a high variation of
the straight line along the image sequence, we
correct it by Double Exponential Smoothing-Based
AUTOMATIC SEGMENTATION OF EMBRYONIC HEART IN TIME-LAPSE FLUORESCENCE MICROSCOPY
IMAGE SEQUENCES
123

Prediction (DESP) (LaViola Jr., 2003) using the
results of the previous images.
Figure 3: The segmented heart (inside line) and the convex
hull (outside line) with convexity defects of the shape
(points).
3.2 Watershed
This method is based on the results of the Watershed
segmentation of subsection 2.5. The general idea is
to divide the segmented shape into the two chambers
by applying a second watershed segmentation.
Therefore, the background is masked out and a
watershed segmentation is applied in this area after a
strong low-pass filtering. If two regions result, then
they correspond to a rough identification of two
chambers. Otherwise the regions have to be joined
until only two regions remain. Therefore, we use the
chamber identification of the previous image. We
compute the intersection of a region in the current
image with the identified chambers of the previous
image. Then, the region is identified to belong to the
chamber where the intersection is maximal. It can
happen that only one region is obtained by the
Watershed segmentation. Then, the segmentation of
the previous image is used for further processing of
the current image.
This chamber identification is very rough
whereas the outline is not coincident with that one of
subsection 2.5 as can be seen in Figure 4. Thus
unassigned pixels remain. In order to assign them to
one of the chambers, an Euclidean distance
transform is computed for each chamber. Then, the
non-assigned pixels of the segmentation are joined
with the chamber for which the distance transform is
smaller.
Figure 4: The Watershed segmentation (outline line) and
the first rough identification of the chambers (inside line).
4 RESULTS AND DISCUSSION
In this section, we show and discuss the results
obtained with the methods presented above. First,
we compare the algorithms for segmenting the shape
of the heart from section 2 using an accuracy
measure. Then, we evaluate visually the results of
chamber segmentation algorithms from section 3.
4.1 Comparison of Segmentation
Algorithms
Several methods exist to measure the performance of
segmentation algorithms (Zhang et al., 2008; Sezgin
and Sankur, 2004). Here, we choose to compare the
segmented images with ground truth images which
were obtained by manual segmentation.
We used the Jaccard coefficient (Cox and Cox,
2001; Ge et al., 2007) as performance measure for
each segmentation method. This coefficient
measures the coincidence between the segmentation
result R and the ground truth A. Then, the
segmentation accuracy is measured as:
,
|
|
|
|
|
|
|
|
|
|
|
|
(2)
with
|
·
|
as the number of pixels of the given region.
The nominator
|
|
means how much of the
object has been detected while the denominator
|
|
is a normalization factor to scale the
accuracy measure into the range of [0,1]. Likewise
pixels falsely detected as belonging to the object
(false positives) are penalized by the normalization
factor. Thus, this accuracy measure is insensitive to
small variations in the ground truth construction and
incorporates both, false positives and negatives, in
one unified function (Ge et al., 2007).
In our experiments we used 26 image sequences
with a resolution of 124×124 pixels. For each image
sequence we segmented the first 20 images with the
above presented methods and compared them with a
ground truth segmentation. We only chose
sequences with fair image quality for evaluation as
otherwise the accuracy of the manual segmentation
is too subjective (Figure 5).
The results of Jaccard coefficient for each sequence
are presented in Table 1. The Voronoi-based and
both thresholding methods outperform the watershed
and level set methods. A visual inspection of the
segmentation results reveals similar results.
BIOSIGNALS 2010 - International Conference on Bio-inspired Systems and Signal Processing
124
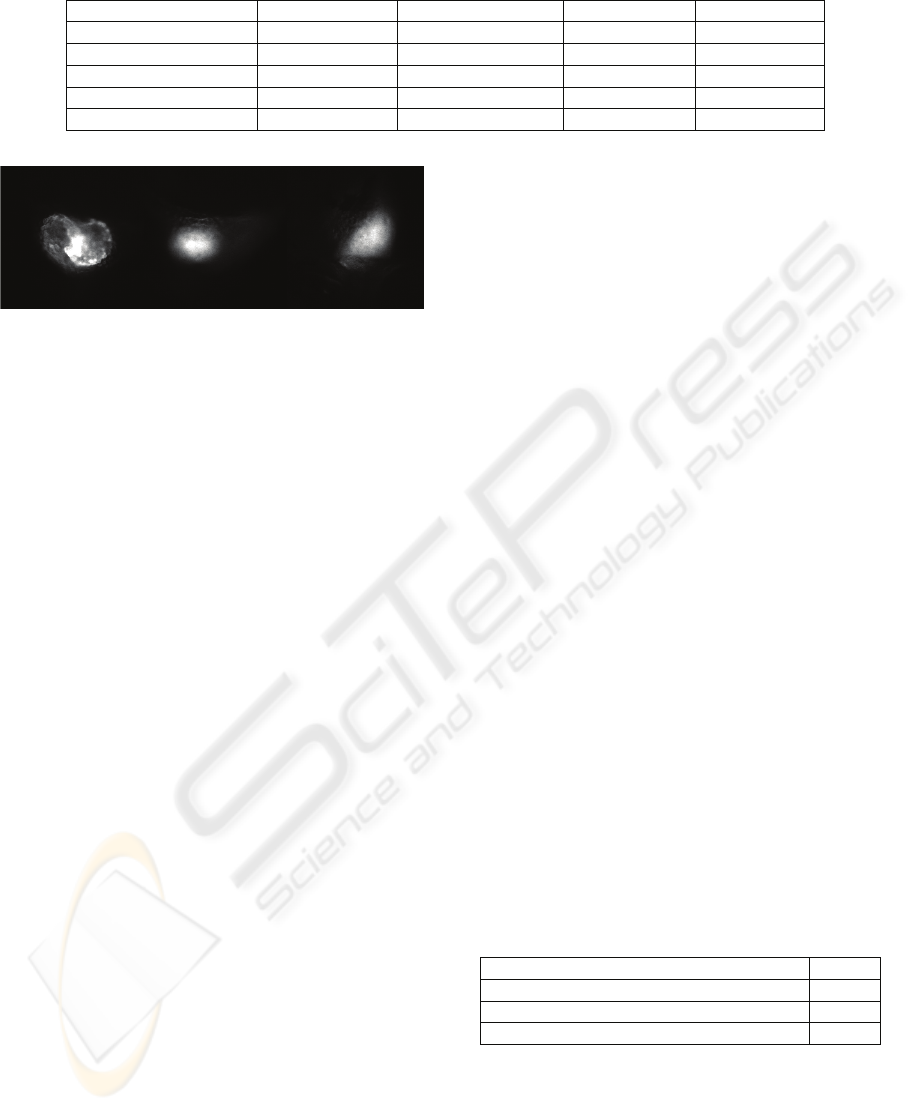
Table 1: Mean accuracy for the segmentation algorithms.
Method Mean accuracy Standard deviation Max accuracy Min accuracy
Adaptive binarization 0.870 0.052 0.946 0.725
Clustering 0.876 0.057 0.954 0.759
Voronoi segmentation 0.890 0.046 0.937 0.769
Level set 0.850 0.062 0.965 0.724
Watershed 0.856 0.044 0.896 0.717
Figure 5: Rejected samples. Overlapping chambers and
blurred images.
The level set method gives good results on high
contrast edges, but in regions where edges are
blurred, the level set does not approach well the
shape of the heart resulting in holes in the object
shape or a too large shape. Moreover, we found it
difficult to determine a common set of parameters
suitable for all sequences.
The contours of the watershed method appear
very rough and are often too tight. This might be due
to the strong low-pass filtering in the post-
processing which causes an edge mismatch. Equally
a false classification of the regions into background
and foreground may cause an inaccurate
segmentation.
The Voronoi-based segmentation method reveals
the best results in term of accuracy measure. The
contours are typically slightly irregular; some
postprocessing could be applied to smooth them. In
case of low-contrast contours it may behave similar
to the level set method. The overall results are quite
satisfying.
The adaptive binarization tends to have a slightly
larger contour, but approaches well the object shape.
This might cause the lower accuracy results, but the
overall segmentation results are good. Sometimes in
case of low-contrast edges the object shape may be
incomplete.
The clustering method tends also to larger
contours, but slightly tighter than the adaptive
binarization method. Therefore, a higher accuracy is
achieved. However, in case of low-contrast edges it
reveals more often incomplete shapes than the
adaptive binarization. Note that the accuracy can
vary as the randomized choice of initial cluster may
result in slightly different segmentation results.
The computational cost cannot be directly
compared as the implementations use different
programming languages and libraries (the adaptive
binarization, clustering, and Voronoi methods are
implemented in C++ using respectively OpenCV,
OpenCV and Torch, and ITK; the level set and
watershed methods are implemented in Matlab).
However, the execution time for each image is
reasonable and estimated at about one second
independently of the method.
4.2 Chamber Identification
In this section, we present some results of the
convexity defects and watershed methods used to
divide the heart into two chambers. The convexity
defects method was evaluated only in combination
with the adaptive binarization and clustering
methods, as they present good segmentation results
(see previous section).
For evaluation we used only 24 out of the 26
sequences from above, because in two other ones the
chambers are superimposed (Figure 5). Such cases
are not taken in consideration in current
developments, and we therefore chose to discard
those sequences. 480 images were then segmented
using each of the described method, and visually
inspected to evaluate whether the heart was correctly
divided. Our results are shown in Table 2, where the
best result is obtained for the adaptive binarization
method.
Table 2: The ratio of correct chamber identification per
image for the chamber identification algorithms.
Method Ratio
Adaptive binarization + convexitydefects 0.704
Clustering + convexity defects 0.577
Watershed 0.456
5 CONCLUSIONS AND FUTURE
WORK
We presented a first attempt of automatically
segmenting the shape and the chambers of the
AUTOMATIC SEGMENTATION OF EMBRYONIC HEART IN TIME-LAPSE FLUORESCENCE MICROSCOPY
IMAGE SEQUENCES
125

zebrafish embryonic heart from time-lapse
fluorescence microscopy image sequences.
For segmenting the shape of the heart, the
Voronoi-based and both thresholding methods
outperform the watershed and level set methods. The
Voronoi-based segmentation gives the best results in
terms of the accuracy measure, as thresholding
methods tend to fail in cases of low-contrast edges.
The watershed segmentation results in quite
rough contours. Anyhow, it is an interesting
approach as it is the basis for chamber identification.
The results of the level set method are not satisfying.
For chamber identification the adaptive binarization
method in combination with the detection of
convexity defects outperforms clearly the other
methods.
Besides segmentation in order to extract
morphological information, we are also working on
other processing methods to extract cardiac function
metrics from image sequence. Such methods are
able to provide additional information for cardiac
development study with very high accuracy.
REFERENCES
Bishop, C.M. (2007). Pattern Recognition and Machine
Learning. (Information Science and Statistics).
Springer.
Cox, T. and Cox, M. (2001). Multidimensional Scaling
(2nd ed.). Chapman & Hall/CRC.
Fink, M., Callol-Massot, C., Chu, A., Ruiz-Lozano, P.,
Belmonte, J. C., Giles, W., Bodmer, R., and Ocorr, K.
(2009). A new method for detection and quantification
of heartbeat parameters in drosophila, zebrafish, and
embryonic mouse hearts. BioTechniques, 46(2), 101–
113.
Ge, F., Wang, S., and Liu, T. (2007). New benchmark for
image segmentation evaluation. Journal of Electronic
Imaging, 16(3):033011.
Hu, N., Sedmera, D., Yost, H., and Clark, E. (2000).
Structure and function of the developing zebrafish
heart. The Anatomical Record, 260(2), 148–157.
Imelinska, C., Downes, M., and Yuan, W. (2002). Semi-
automated color segmentation of anatomical tissue.
Computerized Medical Imaging and Graphics, 24,
173–180.
LaViola Jr., J. (2003). Double exponential smoothing: An
alternative to kalman filter-based predictive tracking.
In Immersive Projection Technology and Virtual
Environments, 199–206.
Li, C., Xu, C., Gui, C., and Fox, M. (2005). Level set
evolution without re-initialization: A new variational
formulation. CVPR, 1, 430–436. IEEE.
Liebling, M., Forouhar, A., Wolleschensky, R.,
Zimmermann, B., Ankerhold, R., Fraser, S., Gharib,
M., and Dickinson, M. E. (2006). Rapid three-
dimensional imaging and analysis of the beating
embryonic heart reveals functional changes during
development. Developmental Dynamics, 235(11),
2940–2948.
Luengo-Oroz, M., Faure, E., Lombardot, B., Sance, R.,
Bourgine, P., Peyri´eras, N., and Santos, A. (2007).
Twister segment morphological filtering. A new
method for live zebrafish embryos confocal images
processing. ICIP, 253–256. IEEE.
Otsu, N. (1979). A threshold selection method from
graylevel histograms. IEEE Trans. on Systems, Man
and Cybernetics, 1(9), 62–69.
Sezgin, M. and Sankur, B. (2004). Survey over image
thresholding techniques and quantitative performance
evaluation. Journal of Electronic Imaging, 13(1), 146–
168.
Vermot, J., Fraser, S., and Liebling, M. (2008). Fast
fluorescence microscopy for imaging the dynamics of
embryonic development. HFSP Journal, 2(3), 143–
155.
Zhang, H., Fritts, J., and Goldman, S. (2008). Image
segmentation evaluation: A survey of unsupervised
methods. Computer Vision and Image Understanding,
110(2), 260–280.
Zuiderveld, K. (1994). Graphics Gems IV, chapter
Contrast Limited Adaptive Histogram Equalization,
pages 474–485. Academic Press.
BIOSIGNALS 2010 - International Conference on Bio-inspired Systems and Signal Processing
126
