
EXPERIMENTS ON SOLVING MULTICLASS RECOGNITION
TASKS IN THE BIOLOGICAL AND MEDICAL DOMAINS
Paolo Soda
Facolt
`
a di Ingegneria, Universit
`
a Campus Bio-Medico di Roma, Via Alvaro del Portillo 21, Roma, Italy
Keywords:
Statistical Pattern Recognition, Decomposition Methods, One-per-class, Reliability Estimation, Classifier En-
sembles.
Abstract:
Multiclass learning problems can be cast as the task of assigning instances to a finite set of classes. Although
in the wide variety of learning tools there exist some algorithms capable of handling polychotomies, many
of the tools were designed by nature for dichotomies. In the literature, many techniques that decompose a
polychotomy into a series of dichotomies have been proposed. One of the possible approaches, known as
one-per-class, is based on a pool of binary modules, where each one distinguishes the elements of one class
from those of the others. In this framework, we propose a novel reconstruction criterion, i.e. a rule that sets the
final decision on the basis of the single binary classifications. It looks at the quality of the current input and,
more specifically, it is a function of the reliability of each classification act provided by the binary modules.
The approach has been tested on four biological and medical datasets and the achieved performance has been
compared with the one previously reported in the literature, showing that the method improves the accuracies
so far.
1 INTRODUCTION
Many supervised pattern recognition tasks can be cast
as the problem of assigning elements to a finite set
of classes or categories. Such tasks are referred to
as binary learning, or dichotomies, when they aim at
distinguishing instances of two classes, whereas they
are named multiclass learning, or polychotomies, if
there are more categories.
There is a huge number of applications that re-
quire multiclass categorization. Some examples are
text classification, object recognition and support to
medical diagnosis, to name a few.
In the literature numerous learning algorithms
have been devised for multiclass problems, such as
neural networks or decision trees. However it ex-
ists a different approach that is based on the reduc-
tion of the multiclass task into multiple binary prob-
lems, referred to as decomposition method. The prob-
lem complexity is therefore reduced trough the de-
composition of the polychotomy in less complex sub-
tasks. The basic observation that supports such an ap-
proach is that in the literature most of the available
algorithms, which handle classification problems, are
best suited to learning binary function (Dietterich and
Bakiri, 1995; Mayoraz and Moreira, 1997). Different
dichotomizers, i.e. the discriminating functions that
subdivide the input patterns in two separated classes,
perform the corresponding recognition task. To pro-
vide the final classification, their outputs are com-
bined according to a given rule, usually referred to
as decision or reconstruction rule.
In the framework of decomposition methods for
classification, the various methods proposed to-date
can be traced back to the following three categories
(Dietterich and Bakiri, 1995; Mayoraz and Moreira,
1997; Jelonek and Stefanowski, 1998; Masulli and
Valentini, 2000; Allwein et al., 2001; Crammer and
Singer, 2002; Hastie and Tibshirani, 1998; Kuncheva,
2005).
The first one, called one-per-class, is based on
a pool of binary learning functions, where each one
separates a single class from all the others. The as-
signment of a new input to a certain class can be
performed, for example, looking at the function that
returns the highest activation (Dietterich and Bakiri,
1995; Masulli and Valentini, 2000).
The second approach, commonly referred to as
distribuited output code, assigns a unique codeword,
i.e. a binary string, to each class. If we assume that
the string has n bit, the recognition system is com-
posed by n binary classification functions. Given
an unknown pattern, the classifiers provide a n-bit
string that is compared with the codeword to set the
64
Soda P. (2008).
EXPERIMENTS ON SOLVING MULTICLASS RECOGNITION TASKS IN THE BIOLOGICAL AND MEDICAL DOMAINS.
In Proceedings of the First International Conference on Bio-inspired Systems and Signal Processing, pages 64-71
DOI: 10.5220/0001066500640071
Copyright
c
SciTePress

final decision. For example, the input sample is as-
signed to the class with the closest codeword, accord-
ing to a distance measure, such as the Hamming one.
In this framework, in (Dietterich and Bakiri, 1995)
the authors proposed an approach, known as error-
correcting techniques (ECOC), where they employed
error-correcting codes as a distributed output repre-
sentation. Their strategy was a decomposition method
based on the coding theory that allowed obtaining a
recognition system less sensitive to noise via the im-
plementation of an error-recovering capability. Al-
though the traditional measure of diversity between
the codewords and the outputs of dichotomizers is
the Hamming distance, other works proposed differ-
ent measures. For example, Kuncheva in (Kuncheva,
2005) presented a measure that accounted for the
overall diversity in the ensemble of binary classifiers.
The last approach is called n
2
classifier. In this
case the recognition system is composed of (n
2
−n)/2
base dichotomizers, where each one is specialized
in discriminating respective pair of decision classes.
Then, their predictions are aggregated to a final deci-
sion using a voting criterion. For example, in (Jelonek
and Stefanowski, 1998) the authors proposed a voting
scheme adjusted by the credibilities of the base classi-
fiers, which were calculated during the learning phase
of the classification.
This short description of the methods so far shows
that the recognition systems based on decomposition
methods are constituted by an ensemble of binary dis-
criminating functions. On this motivation, for brevity
such systems are referred to as Multy Dichotomies
System (MDS) in the following.
In the framework of the one-per-class approach,
we present here a novel reconstruction rule that re-
lies upon the quality of the input pattern and looks
at the reliability of each classification act provided
by the binary modules. Furthermore, the classifica-
tion scheme that we propose allows employing either
a single expert or an ensemble of classifiers internal
to each module that solves a dichotomy. Finally, the
effectiveness of the recognition system has been eval-
uated on four different datasets that belongs to biolog-
ical and medical applications.
The rest of the paper is organized as follows: in
the next section we introduce some notations and
we present general considerations related to the sys-
tem configuration. Section 3 details the reconstruc-
tion method and section 4 describes and discusses
the experiments performed on four different medical
datasets. Finally section 5 offers a conclusion.
2 PROBLEM DEFINITION
2.1 Background
Let us consider a classification task on c data classes,
represented by the set of labels Ω = {ω
1
,· ·· , ω
c
},
with c > 2. With reference to the one-per-class ap-
proach, the multiclass problem is reduced into c bi-
nary problems, each one addressed by one module of
the pool M = {M
1
,· ·· , M
c
}. We say that the module,
or the dichotomizer, M
j
is specialized in the jth class
when it aims at recognizing if the sample x belongs
either to the jth class ω
j
or, alternatively, to any other
class ω
i
, with i 6= j. Therefore each module assigns
to the input pattern x ∈ ℜ
n
a binary label:
M
j
(x) =
1 if x ∈ ω
j
0 if x ∈ ω
i
, i 6= j
(1)
where M
j
(x) indicates the output of the jth module on
the pattern x. On this basis, the codeword associated
to the class ω
j
has a bit equal to 1 at the jth position,
and 0 elsewhere.
Notice that we have just mentioned module and
not classifier to emphasize that each dichotomy can
be solved not only by a single expert, but also by an
ensemble of classifiers. However, to our knowledge,
the system dichotomizers typically adopt the former
approach, i.e. they are composed by one classifier
per specialized module. For example, for their exper-
imental assessments the authors used a a decision tree
and a multi layer perceptrons with one hidden layer
both in (Mayoraz and Moreira, 1997) and (Masulli
and Valentini, 2000), respectively. The same func-
tions were employed by Dietterich and Bakiri for the
evaluation of their proposal in (Dietterich and Bakiri,
1995), whereas Allwein et al. used a Support Vector
Machine (Allwein et al., 2001). A viable alternative to
using a single expert is the combination of classifiers
outputs solving the same recognition task. The idea is
that the classification performance attainable by their
combination should be improved by taking advan-
tage of the strength of the single classifiers. Classi-
fier selection and fusion are the two main combina-
tion strategies reported in the literature. The former
presumes that each classifier has expertise in some
local area of the feature space (Woods et al., 1997;
Kuncheva, 2002; Xu et al., 1992). For example, when
an unknown pattern is submitted for classification, the
more accurate classifier in the vicinity of the input is
selected to label it (Woods et al., 1997). The latter al-
gorithms assume that the classifiers are applied in par-
allel and their outputs are combined to attain some-
how a group of “consensus” (De Stefano et al., 2000;
Kuncheva et al., 2001; Kittler et al., 1998). Typi-
EXPERIMENTS ON SOLVING MULTICLASS RECOGNITION TASKS IN THE BIOLOGICAL AND MEDICAL
DOMAINS
65

cal fusion techniques include weighted mean, voting,
correlation, probability, etc..
It is worth noticing that the modules, besides la-
belling each pattern, may supply other information
typically related to the degree that the sample belongs
to that class. In this respect, the various classifica-
tion algorithms are divided into three categories, on
the basis of the output information that they are able
to provide (Xu et al., 1992). The classifiers of type
1 supply only the label of the presumed class and,
therefore, they are also known as experts that work
at the abstract level. Type 2 classifiers work at the
rank level, i.e. they rank all classes in a queue where
the class at the top is the first choice. Learning func-
tions of type 3 operate at the measurement level, i.e.
they attribute each class a value that measure the de-
gree that the input sample belongs to that class. If
a crisp label of the input pattern is needed, we can
use the maximum membership rule that assigns x to
the class for which the degree of support is maxi-
mum (ties are resolved arbitrarily). Although abstract
classifiers provide a n-bit string that can be compared
with the codewords, decision schemes that exploit in-
formation derived from the classifiers working at the
measurement level permit us to define reconstruction
rules that are potentially more effective. Furthermore,
if the module is constituted by a multi-experts system,
the information supplied by the single classifiers can
be used to compute a measure similar to that provided
by measurement classifiers.
Since measurement classifiers can provide more
information with respect to the other two types, we
assume that only measurement experts constitutes our
MDS. Therefore, the research focus becomes: “Given
the individual decision M
1
(x),· ·· ,M
c
(x) and the de-
grees of membership of x to the different classes, how
can we use such an information to set the final label?”.
2.2 The Reconstruction Method
The reconstruction method addresses the issues of de-
termining the final label of the input pattern x on the
basis of the modules’ decisions and, eventually, of
information directly derived from their outputs. To
present our method, let us introduce two auxiliaries
quantities. The first, named binary profile, represents
the state of the module outputs. It is a c-bit vector
defined by:
M(x) = [M
1
(x),· ·· ,M
j
(x),· ·· ,M
c
(x)] (2)
whose entries are the crisp labels provided by each
module in the classification of sample x (see equa-
tion 1).
Since each block has a binary output, the 2
c
pos-
sible bit combinations of the binary profile can be
grouped into the following three categories:
(i) only one module classifies the sample in the class
in which it is specialized;
(ii) more modules classify the sample in its own
class;
(iii) none module classifies the sample in its own
class.
In the first case, only one entry of M(x) is one; in
the second more elements are one (at least two and no
more than c), whereas in the last situation all the el-
ements are zero. Such an observation naturally leads
to distinguish these three cases using the summation
over the binary profile. Indeed,
m =
c
∑
j=1
M
j
(x) =
1, in case (i)
[2,c], in case (ii)
0, in case (iii)
(3)
where m therefore represents the number of modules
whose outputs are 1.
The second quantity that we introduce is referred
to as reliability profile and it is described by:
ψ(x) = [ψ
1
(x),· ·· ,ψ
j
(x),· ·· ,ψ
c
(x)] (4)
where each element ψ
j
(x) measure the reliability of
the classification act on pattern x provided by the jth
module. Note that the reliability varies in the inter-
val [0, 1], and a value near 1 indicates a very reliable
classification.
We deem that the estimation of the reliability of
each classification act is a viable method to employ
the information directly derived from the classifiers
output since it has demonstrated its convenience, in
other field also (De Stefano et al., 2000; Cordella
et al., 1999).
Assuming that we determined both the binary and
the reliability profiles, i.e. M(x) and ψ(x) respec-
tively, in the next section we will present the recon-
struction rule.
3 RELIABILITY BASED
RECONSTRUCTION
In this section we introduce the novel reconstruction
strategy we propose in the paper. It chooses an output
in any of the 2
c
combinations of the binary profile.
We deem that an accurate final decision can be taken
if the reconstruction rule looks at the quality of the
classification provided by the modules, i.e. at the re-
liability of their specific decisions. To our knowledge
the application of such a parameter can not be found
in the literature related to decomposition methods. In-
deed, the papers of this field that used the information
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
66

directly derived from the outputs of the base classi-
fiers typically considered only the highest activation
among the experts, e.g. the maximum output from a
pool of neural networks. However, this measure can-
not be regarded as a reliability parameter, since it has
been demonstrated that it should be computed consid-
ering not only the winner output neurons but also the
losers (Cordella et al., 1999).
Therefore, differently from the past, we propose a
criterion that makes use of the reliability measure, i.e.
of the reliability profile, named as Reliability-based-
Reconstruction (RbR). Denoting by s the index of the
module that sets the final output O(x) ∈ Ω, referred
to as selected module for brevity in the following, the
final decision is given by:
O(x) = ω
s
(5)
with
s =
argmax
j
(M
j
(x) · ψ
j
(x)), if m ∈ [1,c]
argmin
j
(M
j
(x) · ψ
j
(x)), if m = 0
(6)
where M
j
(x) indicates the negate output of the jth
block.
The first row of this equation considers both cases
(i) and (ii). Indeed, since in the first case all the mod-
ules agree in their decision, as a final output is chosen
the class of the module whose output is 1. Conversely,
in cases (ii) and (iii) the final decision is performed
looking at the reliability of each modules’ classifica-
tions. In case (ii), m modules vote for their own class,
whereas the others (c − m) indicate that x does not
belong to their own class. To solve the dichotomy
between the m conflicting modules we look at the re-
liability of their classification and choose the class as-
sociated to the more reliable one. In case (iii) m = 0,
suggesting that all modules classify x as belonging to
another class than the one they are specialized. In this
case, the bigger is the reliability parameter ψ
j
(x), the
less is the probability that x belongs to ω
j
, and the
bigger is the probability that it belongs to the other
classes. These observations suggest finding out which
module has the minimum reliability and then choos-
ing the class associated to it as a final output.
Panel A of figure 1 shows the architecture of the
proposed recognition system. The decision M
j
(x) and
the reliability ψ
j
(x) supplied by each of the c mod-
ules are aggregated in the reconstruction module to
provide the final decision O(x). As observed in sec-
tion 2.1, the use of an ensemble of classifiers in each
module is a way to improve its discrimination capa-
bility. In this respect, the panel B of the same figure
depicts a typical configuration of a multi-experts sys-
tem. Notice that both the output of the kth classifier
and its reliability, denoted as V
k
(x) and ξ
k
(x), respec-
tively, can be given to the combination rule in order
to label the input sample.
4 EXPERIMENTAL EVALUATION
In this section we first describe the datasets used to
assess the performance of the reconstruction method
and, second, we briefly discuss the configuration of
the MDS modules. Third, we present a strategy to
evaluate the classification reliability when the mod-
ules are constituted both by a single classifier and by
an ensemble of experts, respectively. Finally, we re-
port the experimental results.
4.1 Datasets
For our tests we use four datasets, described in the
following and summerized in table 1.
Indirect Immunofluorescence Well Fluores-
cence Intensity. Connective tissue diseases are
autoimmune disorders characterized by a chronic
inflammatory process involving connective tissues.
When they are suspected in a patient, the Indirect Im-
munofluorescence (IIF) test based on HEp-2 substrate
is usually performed, since it is the recommended
method. The interested reader may find a wide expla-
nation of the IIF and its issues in (Kavanaugh et al.,
2000; Rigon et al., 2007). The dataset consists of 14
features extracted from 600 patients sera collected
at Universit
`
a Campus Bio-Medico di Roma. The
samples are distributed over three classes, namely
positive (36.0%), negative (32.5%) and intermediate
(31.5%). Previous results are reported in (Soda
and Iannello, 2006) where the authors employed a
multiclass approach, achieving an accuracy of 76%
approximately.
Indirect Immunofluorescence HEp-2 cells stain-
ing pattern. This is a dataset with 573 instances
represented by 159 statistical and spectral features.
The samples are distributed in five classes that are
representative of the main staining patterns exhibited
by HEp-2 cells, namely homogeneous (23.9%),
peripheral nuclear (21.8%), speckled (37.0%), nu-
cleolar (8.2%) and artefact (9.1%). These patterns
are related to one of the different autoantibodies
that give rise to a connective tissue disease. For
the details on these classes see (Rigon et al., 2007).
On this dataset, we performed some tests adopting
a multiclass approach, which exhibits a hit rate of
63.6% approximately, evaluated using the eightfold
cross validation.
EXPERIMENTS ON SOLVING MULTICLASS RECOGNITION TASKS IN THE BIOLOGICAL AND MEDICAL
DOMAINS
67
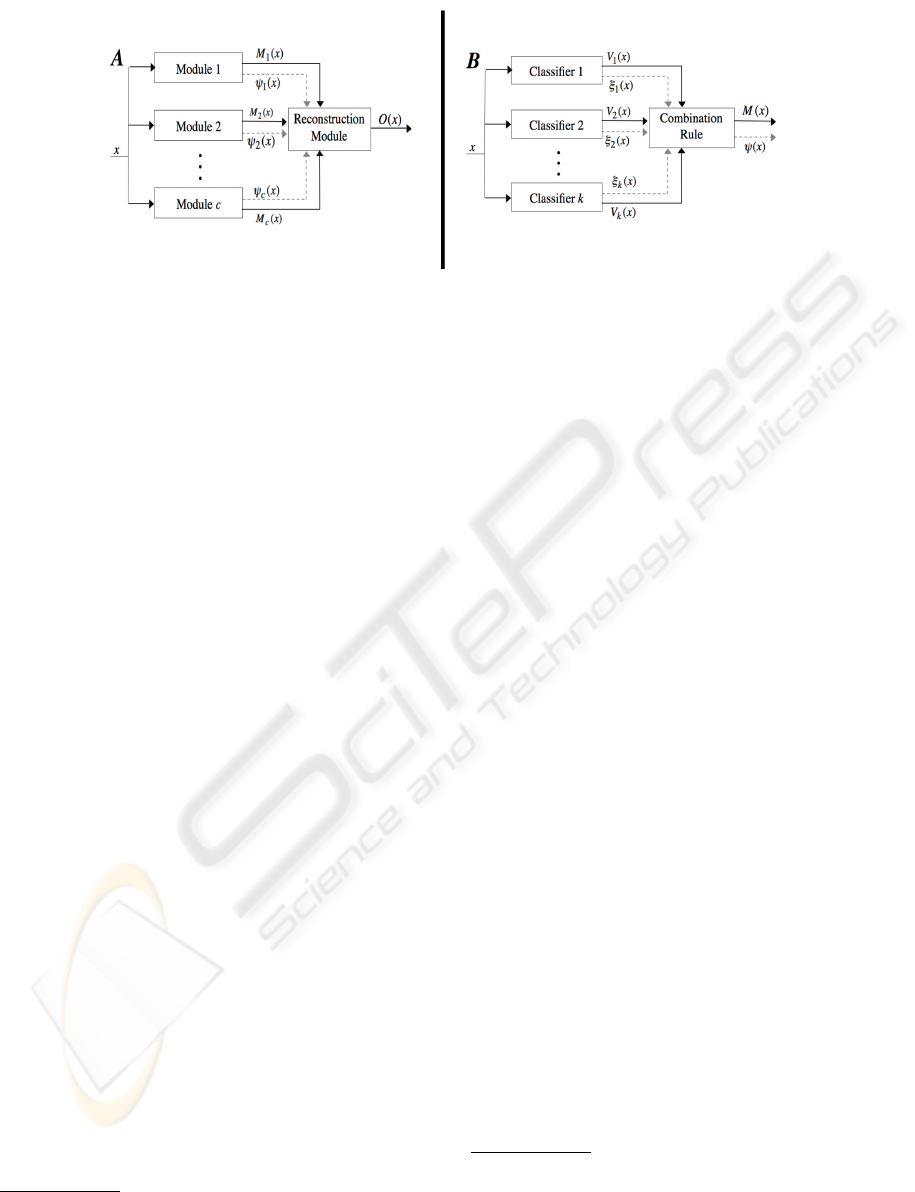
Figure 1: The system architecture, which is based on the aggregation of binary modules (panel A), according to the one-per-
class approach. Note that each module can be constituted by a multi-experts system, as depicted in the panel B.
Lymphography. A database of lymph diseases
was obtained from the University Medical Centre,
Institute of Oncology, Ljubljana. It is composed by
148 instances described by 18 numeric attributes.
There are four classes, namely normal (1.4%),
metastases (54.7%), malign lymph (41.2%) and
fibrosis (2.7%). The data are available within the
UCI Machine Learning Repository
1
(Asuncion and
Newman, 2007). Different approaches were used
in the literature to address the recognition task.
For instance, for Naive Bayes classifier and C4.5
decision tree the achieved performance was 79% and
77% respectively (Clark and Niblett, 1987), whereas
induction techniques correctly classified the 83% of
samples (Cheung, 2001).
Ecoli. The database is composed by 336 sam-
ples, described by a nine-dimensional vector and
distributed in eight classes. Each class represents a
localization site, which can be cytoplasm (42.5%),
inner membrane without signal sequence (22.9%),
periplasm (15.5%), inner membrane, uncleavable
signal sequence (10.4%), outer membrane (6.0%),
outer membrane lipoprotein (1.5%), inner membrane
lipoprotein (0.6%) and inner membrane, cleavable
signal sequence (0.6%). Again, the data are avail-
able within the UCI Machine Learning Repository
(Asuncion and Newman, 2007). In (Jelonek and
Stefanowski, 1998), the authors reported an accuracy
that ranges from 79.7% up to 83.0%, achieved
employing both a decision tree and a Multi Layer
Perceptrons, respectively. In (Allwein et al., 2001),
using many popular classification algorithms, such as
the support-vector machines, AdaBoost, regression
1
For each dataset of this repository the users have access
to a description of the application domain, to the features
and to the ground truth.
and decision-tree algorithms, the hit rate varies from
78.5% up to 86.1%.
4.2 MDS Configuration
The modules of the MDS are essentially composed by
a single classifier or by an ensemble of classifiers. In
both cases, as single expert we use k-Nearest Neigh-
bour (kNN) or Multi-Layer Perceptron (MLP). For
each dichotomy, we first select a subset of features
that simplifies both the pattern representation and the
classifier complexity as well as the risk of the incur-
ring in the peaking phenomenon
2
. Then we carry out
some preliminary tests to determine the best config-
uration of experts parameters, e.g. the number of
neighbours for kNN classifier or the number of hidden
layers, neurons per layer, etc., for the MLP network.
Furthermore, when the module is constituted by an
ensemble of experts we adopt a fusion technique to
combine their outputs, namely the Weighted Voting
(WV). In such a method the opinion of each expert
about the class of the input pattern is weighted by
the reliability of its classification. Since each expert
deals with a binary learning task, to further present
this scheme we can simplify the notation as follows.
Denoting as V
k
(x) and as ξ
k
(x) the output and the clas-
sification reliability of kth classifier on sample x, the
weighted sum of the votes for each of the two classes
is given by:
W
h
(x) =
∑
k:V
k
(x)=h
ξ
k
(x), with h = {0,1} (7)
2
The peaking phenomenon is a paradoxical behaviour
in which the added features may actually degrade the per-
formance of a classifier if the number of training samples
that are used to design the classifier is small relative to the
number of features.
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
68
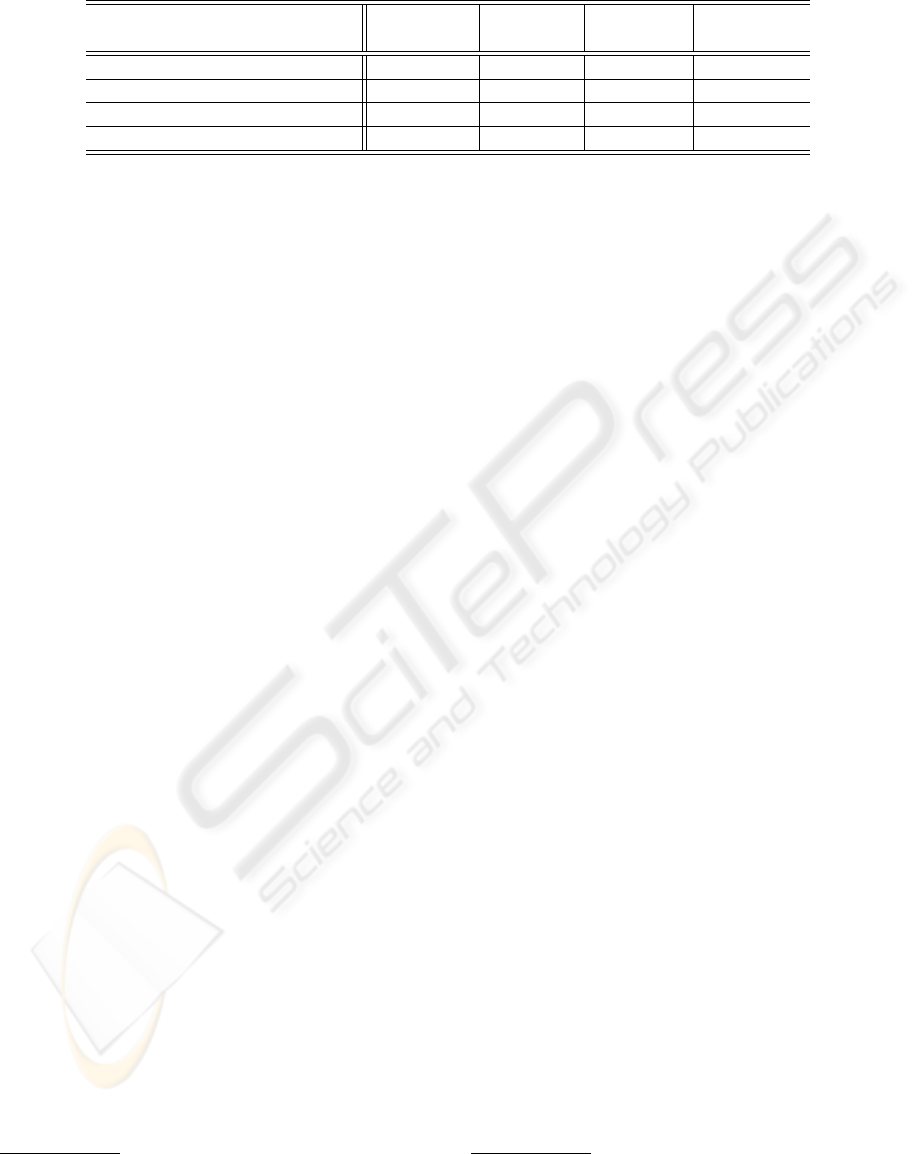
Table 1: Summary of the datasets used.
Database Number Number Number Avalaibility
of Samples of Classes of features
IIF Well Fluorescence Intensity 600 3 14 Private
IIF HEp-2 cells staining pattern 573 5 159 Private
Lymphography 148 4 18 UCI
Ecoli 336 8 9 UCI
The output of the fusion of the jth module, M
j
(x), is
defined by
3
:
M
j
(x) =
1 if W
1
(x) > W
0
(x)
0 otherwise
(8)
Turning our attention to the configuration of the
system in the experimental tests, notice that the mod-
ules that label the samples of the IIF Well Fluo-
rescence Intensity and of lymphography datasets are
composed by one classifiers. The modules that clas-
sify the samples of the HEp-2 cells and of the ecoli
databases are constituted by kNN and MLP classifiers
combined by the WV criterion.
4.3 Reliability Parameter
The approach described for deriving the final decision
according to the RbR rule requires the introduction
of a reliability parameter that evaluates the quality of
the classification performed by each module, which
can be composed by a single classifier or by an ag-
gregation of classifiers (figure 1). In the former case
its reliability ψ
j
coincides with the one of the single
classifier, i.e. ξ. In the latter case, each entry of the
reliability profile generally depends on the combina-
tion rule adopted in the module, on the number k of
composing experts and on their individual reliabilities
ξ. Formally,
ψ
j
(x) =
ξ(x), if k = 1
f (ξ
1
(x),· ·· , ξ
i
(x),· ·· , ξ
k
(x)), if k > 1
(9)
where all the reliabilities are reported as function of
the input pattern to emphasize that they are computed
for each classification act.
In the rest of this section we first present two tech-
niques to measure the reliability of kNN and MLP de-
cisions, and then we introduce a novel method that es-
timates such parameter in the case of the application
of the WV criterion.
A typical approach that measures the reliability of
the decision taken by the single expert, i.e. ξ, makes
3
In case of tie, i.e. if W
1
(x) is equal to W
0
(x), the output
M
j
(x) is set arbitrarily to zero. Note that it never occurred
in all tests we performed.
use of the confusion matrix
4
estimated on the learn-
ing set. The drawback of this method is that all the
patterns with the same label have equal reliability, re-
gardless of the quality of the sample. Indeed, the aver-
age performance on the learning set, although signifi-
cant, does not necessarily reflect the actual reliability
of each classification act. To overcome such limita-
tions we adopt an approach that relies upon the quality
of the current input. To this end, we refer to the work
presented in (Cordella et al., 1999), where the quality
of the sample is related to its position in the feature
space. In this respect, the low reliability of a recog-
nition act can be traced back to one of the following
situations: (a) in the feature space x is located in a re-
gion that is far from those associated with the various
classes, i.e. the sample is significantly different from
those present in the training set, (b) the point repre-
senting x lies in a region of the feature space where
two or more classes overlap. These observations lead
to introduce the parameters ξ
a
and ξ
b
that distinguish
between the two situations of unreliable classification.
Then, a comprehensive parameter ξ can be derived
adopting the following conservative choice:
ξ = min(ξ
a
,ξ
b
) (10)
Indeed, it implies that a low value for only one of
the parameters is sufficient to consider unreliable the
classification.
In the case of kNN classifiers, following (Cordella
et al., 1999), the two parameters are defined are given
by:
ξ
a
= max (1 − D
min
/D
max
,0) (11)
ξ
b
= 1 − D
min
/D
min2
(12)
where D
min
is the smallest distance of x from a ref-
erence sample belonging to the same class of x, D
max
is the highest among the values of D
min
obtained for
samples taken from the training-test set, i.e. a set that
is disjoint from both the reference and the test set,
D
min2
is the distance between x and the reference sam-
ple with the second smallest distance from x among
4
The confusion matrix reports for each entry (p,q) the
percentage of samples of the class C
p
assigned to the class
C
q
.
EXPERIMENTS ON SOLVING MULTICLASS RECOGNITION TASKS IN THE BIOLOGICAL AND MEDICAL
DOMAINS
69

all the reference set samples belonging to a class that
is different from that determining D
min
.
In the case of MLP classifier, the two quantities
are defined as follows:
ξ
a
= N
win
(13)
ξ
b
= N
win
− N
2win
(14)
where N
win
is the output of the winner neuron, N
2win
is the output of the neuron with the highest value after
the winner. From this definition, it is straightforward
that ξ = ξ
b
. For further details see (De Stefano et al.,
2000).
When the jth module is composed by more than
one classifier combined according to the WV rule,
the reliability estimator considers again the situations
which can give rise to an unreliable classification. In
this respect, we need to introduce the following two
auxiliary quantities:
π
1
(x) = max ({ξ
k
(x)|k : V
k
(x) = M
j
(x)}) (15)
π
2
(x) = max ({ξ
k
(x)|k : V
k
(x) 6= M
j
(x)} ∪ {0})
(16)
where π
1
(x) and π
2
(x) represent the maximum relia-
bilities of experts voting for the winning class and for
other classes (0 if all the experts agree on the winner
class), respectively. Given these definitions, the reli-
ability of the WV rule can be evaluated according to
the following conservative choice:
ψ(x) = min (π
1
(x),max (0,1 − π
2
(x)/π
1
(x))) (17)
4.4 Results and Discussion
This section presents the experimental results that we
achieved using the system described so far. To evalu-
ate and then compare the results of this approach with
those reported in the literature we perform eightfold
and tenfold cross validation on the two IIF datasets,
i.e. well fluorescence intensity and HEp-2 cells stain-
ing pattern, and on the other two databases, i.e. lym-
phography and ecoli, respectively.
The third column of table 2 shows the testing ac-
curacies achieved on the four databases. To sim-
ply compare them with the past results, the second
column of the same table summarizes the perfor-
mance reported in literature. Turning our attention
to the tests carried out on the first and on the sec-
ond datasets, a relevant accuracy improvement can
be observed. Indeed, the hit rate increases of 18.4%
and of 12.3% in the case of the well fluorescence in-
tensity and HEp-2 cells staining pattern databases,
respectively. In our opinion, such an improvement
is twofold motivated. On the one hand, the set of
extracted features is more stable and more effective
when we adopt a decomposition approach rather than
a multiclass one. On the other hand, the reconstruc-
tion rule exhibits a very good capability of solving
the disagreements between the specialized modules.
Indeed, when the binary profile of the input sample
M(x) differs from one of the possible codewords (i.e.
m = 0 or 2 ≤ m ≤ c), the decision is taken looking
at the reliability profile ψ(x), as presented in the for-
mula 6. These considerations are strengthened by
the observation of the performance attained in the
classification of samples belonging to the two UCI
datasets. Indeed, since they are benchmark datasets,
any reported improvement is due to the recognition
approach rather than to the use of a different features
set. The tests on both the lymphography and ecoli
datasets exhibit an accuracy better than the one re-
ported to date. Indeed, for the former dataset the
improvement ranges both from 6.9% up to 12.9% ,
whereas for the latter one it varies from 1.8% up to
9.4%. Therefore, also in these cases the MDS in
combination with the RbR rule improves the recog-
nition performance. Furthermore, it is worth noting
that the approach seems independent of the modules’
arrangement. The rationale lies in observing that in
two of the four tests the MDS modules are consti-
tuted by a multi-experts system, whereas in the others
they are composed by a single classifier (see the be-
ginning of section 4). Consequently, the reliability ψ
j
is measured according to a method that varies with the
module configuration, as previously presented (see
equations 10-17). Nevertheless, these variations do
not affect the effectiveness of the recognition system.
Therefore, we deem that the reconstruction rule is ro-
bust with respect to different reliability estimators.
5 CONCLUSIONS
In the framework of decomposition methods, we have
presented a classification approach that reconstructs
the final decision looking at the reliability of each
classification act provided by all dichotomizers. Fur-
thermore, the reconstruction rule does not depend on
the configuration of each module, i.e. on its archi-
tecture. Such an observation is strengthened by the
good performance achieved when both a single clas-
sifier and a fusion of experts constitute each module,
respectively.
For all the four tested databases, the experimental
results show that the proposed system outperforms the
performance reported in the literature.
Future works are directed towards two issues.
First, the test of the system on other public datasets
and, second, the definition of reliability parameter of
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
70

Table 2: Testing accuracy achieved on the used datasets.
Database Past MDS using
Usage RbR
IIF Well Fluorescence Intensity 75.9% 94.3%
IIF HEp-2 cells staining pattern 63.6% 75.9%
Lymphography 77% − 83.0% 89.9%
Ecoli 78.5% − 86.1% 87.9%
each decision taken by the MDS.
ACKNOWLEDGEMENTS
The author would like to thank the DAS s.r.l (www.
dasitaly.com), which has funded this work.
REFERENCES
Allwein, E. L., Schapire, R. E., and Singer, Y. (2001). Re-
ducing multiclass to binary: a unifying approach for
margin classifiers. J. Mach. Learn. Res., 1:113–141.
Asuncion, A. and Newman, D. J. (2007). UCI machine
learning repository.
Cheung, N. (2001). Machine learning techniques for med-
ical analysis. Master’s thesis, University of Queens-
land.
Clark, P. and Niblett, T. (1987). Induction in noisy domains.
In Progress in Machine Learning–Proc. of EWSL 87,
pages 11–30.
Cordella, L., Foggia, P., and et. al. (1999). Reliability pa-
rameters to improve combination strategies in multi-
expert systems. Patt. An. & Appl., 2(3):205–214.
Crammer, K. and Singer, Y. (2002). On the algorithmic
implementation of multiclass kernel-based vector ma-
chines. J. Mach. Learn. Res., 2:265–292.
De Stefano, C., Sansone, C., and Vento, M. (2000). To
reject or not to reject: that is the question: an answer
in case of neural classifiers. IEEE Trans. on Systems,
Man, and Cybernetics–Part C, 30(1):84–93.
Dietterich, T. G. and Bakiri, G. (1995). Solving multiclass
learning problems via error-correcting output codes.
Journal of Artificial Intelligence Research, 2:263.
Hastie, T. and Tibshirani, R. (1998). Classification by pair-
wise coupling. In NIPS ’97: Proc. of the 1997 Conf.
on Advances in neural information processing sys-
tems, pages 507–513. MIT Press.
Jelonek, J. and Stefanowski, J. (1998). Experiments on
solving multiclass learning problems by n
2
classifier.
In 10th European Conference on Machine Learning,
pages 172–177. Springer-Verlag Lecture Notes in Ar-
tificial Intelligence.
Kavanaugh, A., Tomar, R., and et al. (2000). Guidelines for
clinical use of the antinuclear antibody test and tests
for specific autoantibodies to nuclear antigens. Am.
Col. of Pathologists, Archives of Pathology and Lab.
Medicine, 124(1):71–81.
Kittler, J., Hatef, M., and et. al. (1998). On combining clas-
sifiers. IEEE Trans. On Pattern Analysis and Machine
Intelligence, 20(3):226–239.
Kuncheva, L. I. (2002). Switching between selection and
fusion in combining classifiers: an experiment. IEEE
Trans. on Systems, Man and Cybernetics, Part B,
32(2):146–156.
Kuncheva, L. I. (2005). Using diversity measures for gen-
erating error-correcting output codes in classifier en-
sembles. Patt. Recogn. Lett., 26(1):83–90.
Kuncheva, L. I., Bezdek, J. C., and Duin, R. (2001). Deci-
sion template for multiple classifier fusion: an experi-
mental comparison. Patt. Recognition, 34:299–314.
Masulli, F. and Valentini, G. (2000). Comparing decompo-
sition methods for classication. In KES’2000, Fourth
Int. Conf. on Knowledge-Based Intell. Eng. Systems &
Allied Technologies, pages 788–791.
Mayoraz, E. and Moreira, M. (1997). On the decompo-
sition of polychotomies into dichotomies. In ICML
’97: Proc. of the 14th Int. Conf. on Machine Learning,
pages 219–226. Morgan Kaufmann Publishers Inc.
Rigon, A., Soda, P., Zennaro, D., Iannello, G., and Afeltra,
A. (2007). Indirect immunofluorescence (IIF) in au-
toimmune diseases: Assessment of digital images for
diagnostic purpose. Cytometry - Accepted for Publi-
cation, February.
Soda, P. and Iannello, G. (2006). A multi-expert system to
classify fluorescent intensity in antinuclear autoanti-
bodies testing. In Computer Based Medical Systems,
pages 219–224. IEEE Computer Society.
Woods, K., Kegelmeyer, W., and Bowyer, K. (1997). Com-
bination of multiple classifiers using local accuracy
estimates. IEEE Transactions on Pattern Analysis and
Machine Intelligence, 19:405–410.
Xu, L., Krzyzak, A., and Suen, C. (1992). Method of
combining multiple classifiers and their application to
handwritten numeral recognition. IEEE Trans. on Sys-
tems, Man and Cybernetics, 22(3):418–435.
EXPERIMENTS ON SOLVING MULTICLASS RECOGNITION TASKS IN THE BIOLOGICAL AND MEDICAL
DOMAINS
71
