
EVOLUTIONARY COMPUTATION APPROACH TO
ECG SIGNAL CLASSIFICATION
Farid Melgani
Dept. of Information and Communicatrion technologies, University of Trento, Via Sommarive, 14-I-38050, Trento, Italy
Yakoub Bazi
College of Engineering, Al Jouf University, 2014 Sakaka, Saudi Arabia
Keywords: ECG classification, feature reduction, particle swarm optimization, support vector machine.
Abstract: In this paper, we propose a novel classification system for ECG signals based on particle swarm
optimization (PSO). The main objective of this system is to optimize the performance of the support vector
machine (SVM) classifier in terms of accuracy by automatically: i) searching for the best subset of features
where to carry out the classification task; and ii) solving the SVM model selection issue. Experiments
conducted on the basis of ECG data from the MIT-BIH arrhythmia database to classify five kinds of
abnormal waveforms and normal beats confirm the effectiveness of the proposed PSO-SVM classification
system.
1 INTRODUCTION
The recent literature reports different and interesting
methodologies for the automatic classification of
electrocardiogram (ECG) signals (e.g., de Chazal et
Reilly, 2006, and Inan et Giovangrandi, 2006).
However, in the design of an ECG classification
system, there are still some open issues, which if
suitably addressed may lead to the development of
more robust and efficient classifiers. One of these
issues is related to the choice of the classification
approach to be adopted. In particular, we think that,
despite its great potential, the SVM approach has not
received the attention it deserves in the ECG
classification literature compared to other research
fields. Indeed, the SVM classifier exhibits a
promising generalization capability thanks to the
maximal margin principle (MMP) it is based on
(Vapnik, 1998). Another important property is its
lower sensitivity to the curse of dimensionality
compared to traditional classification approaches.
This is explained by the fact that the MMP makes
unnecessary to estimate explicitly the statistical
distributions of classes in the hyperdimensional
feature space in order to carry out the classification
task. Thanks to these interesting properties, the SVM
classifier proved successful in numerous and
different application fields, such as 3D object
recognition (Pontil et Verri, 1998), biomedical
imaging (El-Naqa et al., 2002), remote sensing
(Melgani et Bruzzone, 2004 and Bazi et Melgani,
2006). Turning back to ECG classification, other
issues which can be identified are: 1) feature
selection is not performed in a completely automatic
way; and 2) the selection of the best free parameters
of the adopted classifier is generally performed
empirically (model selection issue).
In this paper, we present in a first step a thorough
experimental exploration of the SVM capabilities for
ECG classification. In a second step, in order to
address the aforementioned issues, we propose to
optimize further the performances of the SVM
approach in terms of classification accuracy by 1)
automatically detecting the best discriminating
features from the whole considered feature space
and 2) solving the model selection issue. The
detection process is implemented through a particle
swarm optimization (PSO) framework that exploits a
criterion intrinsically related to the SVM classifier
properties, namely the number of support vectors
(#SV).
19
Melgani F. and Bazi Y. (2008).
EVOLUTIONARY COMPUTATION APPROACH TO ECG SIGNAL CLASSIFICATION.
In Proceedings of the First International Conference on Bio-inspired Systems and Signal Processing, pages 19-24
DOI: 10.5220/0001060200190024
Copyright
c
SciTePress

2 PROPOSED APPROACH
2.1 Support Vector Machine (SVM)
Let us first for simplicity consider a supervised
binary classification problem. Let us assume that the
training set consists of N vectors x
i
∈ ℜ
d
(i = 1, 2,
…, N) from the d-dimensional feature space X. To
each vector x
i
, we associate a target y
i
∈ {-1, +1}.
The linear SVM classification approach consists of
looking for a separation between the two classes in
X by means of an optimal hyperplane that
maximizes the separating margin (Vapnik, 1998). In
the nonlinear case, which is the most commonly
used as data are often linearly nonseparable, they are
first mapped with a kernel method in a higher
dimensional feature space, i.e., Φ(X) ∈ ℜ
d’
(d’> d).
The membership decision rule is based on the
function sign[f(x)], where f(x) represents the
discriminant function associated with the hyperplane
in the transformed space and is defined as:
f(x) = w
*
⋅Φ(x) + b
*
(1)
The optimal hyperplane defined by the weight
vector w
*
∈
ℜ
d’
and the bias b
*
∈
ℜ
is the one that
minimizes a cost function that expresses a
combination of two criteria: margin maximization
and error minimization. It is expressed as:
∑
=
+=
N
i
i
ξC
1
2
1
)(
2
wξw,Ψ
(2)
This cost function minimization is subject to the
following constraints:
i
ξ1b
i
Φ
i
y
−
≥+⋅ ))(( xw , i = 1,…, N
(3)
and
0≥
i
ξ , i = 1, 2, …, N
(4)
where the
ξ
i
’s are slack variables introduced to
account for non-separable data. The constant C
represents a regularization parameter that allows to
control the shape of the discriminant function. The
above optimization problem can be reformulated
through a Lagrange functional, for which the
Lagrange multipliers can be found by means of a
dual optimization leading to a Quadratic
Programming (QP) solution, i.e.,
∑∑
==
−
N
1i
N
ji,
jijijii
,Kyyααα
1
)(
2
1
max xx
α
(5)
under the constraints:
0≥
i
α
for i = 1, 2, …, N
(6)
and
∑
=
=
N
1i
ii
0yα
(7)
where α=[α
1
, α
2
,…, α
N
] is the vector of Lagrange
multipliers and
)(
⋅
⋅
,K is a kernel function. The final
result is a discriminant function conveniently
expressed as a function of the data in the original
(lower) dimensional feature space X:
∑
∈
+=
Si
*
ii
*
i
b,Kyα)f( )( xxx
(8)
The set S is a subset of the indices {1, 2, …, N}
corresponding to the non-zero Lagrange multipliers
α
i
’s, which define the so-called support vectors.
As described above, SVMs are intrinsically binary
classifiers. But the classification of ECG signals
often involves the simultaneous discrimination of
numerous information classes. In order to face this
issue, different multiclass classification strategies
can be adopted (Melgani et Bruzzone, 2004). In this
paper, we shall consider the commonly used one-
against-all strategy.
2.2 PSO Principles
Particle swarm optimization (PSO) is a stochastic
optimization technique introduced recently by
Kennedy and Eberhart, which is inspired by social
behavior of bird flocking and fish schooling
(Kennedy et Eberhart, 2001). Similarly to other
evolutionary computation algorithms such as genetic
algorithms (GAs) (Bazi et Melgani, 2006), PSO is a
population-based search method, which exploits the
concept of social sharing of information. This means
that each individual (called particle) of a given
population (called swarm) can profit from the
previous experiences of all other individuals from
the same population. During the iterative search
process in the d-dimensional solution space, each
particle (i.e., candidate solution) will adjust its flying
velocity and position according to its own flying
experience as well as the experiences of the other
companion particles of the swarm.
Let us consider a swarm of size S. Each
particle
) ,...,2 ,1( SiP
i
=
from the swarm is
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
20

characterized by: 1) its current position
d
i
t ℜ∈)(p ,
which refers to a candidate solution of the
optimization problem at iteration t; 2) its velocity
d
i
t ℜ∈)(v ; and 3) the best position
d
bi
t ℜ∈)(p
identified during its past trajectory. Let
d
g
t ℜ∈)(p
be the best global position found over all trajectories
traveled by the particles of the swarm. The position
optimality is measured by means of one or more
fitness functions defined in relation to the considered
optimization problem. During the search process, the
particles move according to the following equations:
()
()
)()()(
)()()()()1(
22
11
tttrc
tttrctwt
ig
ibiii
pp
ppvv
−⋅+
−⋅+=+
)()()1( ttt
iii
vpp +=+
(9)
(10)
where r
1
(⋅) and r
2
(⋅) are random variables drawn
from a uniform distribution in the range [0, 1] so that
to provide a stochastic weighting of the different
components participating in the particle velocity
definition. c
1
and c
2
are two acceleration constants
regulating the relative velocities with respect to the
best global and local positions, respectively. The
inertia weight w is used as a tradeoff between global
and local exploration capabilities of the swarm.
2.3 PSO Setup
The position
2+
ℜ∈
d
i
p of each particle P
i
from the
swarm is viewed as a vector encoding: 1) a
candidate subset F of features among the d available
input features, and 2) the value of the two SVM
classifier parameters, which are the regularization
and the kernel parameters C and γ, respectively.
Since the first part of the position vector implements
a feature detection task, each component
(coordinate) of this part will assume either a “0”
(feature discarded) or a “1” (feature selected) value.
The conversion from real to binary values will be
done by a simple thresholding operation at the 0.5
value.
Let f(i) be the fitness function value associated
with the ith particle P
i
. The choice of the fitness
function is important since, on its basis, the PSO
evaluates the goodness of each candidate solution p
i
for designing our SVM classification system. A
possible choice is to adopt the class of criteria that
estimates the leave-one-out error bound, which
exhibits the interesting property of representing an
unbiased estimation of the generalization
performance of classifiers. In particular, for SVM
classifiers, different measures of this error bound
have been derived, such as the radius-margin bound
and the simple support vector (SV) count (Vapnik,
1998). In this paper, we will explore the simple SV
count as fitness criterion in the PSO optimization
framework because of its simplicity and
effectiveness as shown in the context of the
classification of hyperspectral remote sensing
images (Bazi et Melgani, 2006).
2.4 SVM Classification with PSO
• Initialization
Step 1: Generate randomly an initial swarm of size
S.
Step 2: Set to zero the velocity vectors v
i
(i= 1, 2,...,
S) associated with the S particles.
Step 3: For each position
2+
ℜ∈
d
i
p of the particle
) ,...,2 ,1( SiP
i
=
from the swarm, train an SVM
classifier and compute the corresponding fitness
function f(i) (i.e., the #SV measure).
Step 4: Set the best position of each particle with its
initial position, i.e.,
ibi
pp
=
, (i=1, 2,.., S)
(11)
• Search process
Step 5: Detect the best global position
g
p in the
swarm exhibiting the minimal value of the
considered fitness function over all explored
trajectories.
Step 6: Update the speed of each particle using
Equation (9).
Step 7: Update the position of each particle using
Equation (10). If a particle goes beyond the
predefined boundaries of the search space, truncate
the updating by setting the position of the particle at
the space boundary and reverse its search direction
(i.e., multiply its speed vector by -1). This will
permit to forbid the particles from further attempting
to go outside the allowed search space.
Step 8: For each candidate particle p
i
(i= 1, 2,..., S),
train an SVM classifier and compute the
corresponding fitness function.
Step 9: Update the best position
bi
p of each particle
if its new current position
i
p (i= 1, 2,..., S) has a
smaller fitness function.
• Convergence
Step 10: If the maximal number of iterations is not
yet reached, return to Step 5.
• Classification
Step 11: Select the best global position
*
g
p in the
swarm and train an SVM classifier fed with the
subset of detected features mapped by
*
g
p
and
modeled with the values of the two parameters C
and γ encoded in the same position.
Step 12: Classify the ECG signals with the trained
SVM classifier.
EVOLUTIONARY COMPUTATION APPROACH TO ECG SIGNAL CLASSIFICATION
21

3 EXPERIMENTAL RESULTS
Our experiments were conducted on the basis of
ECG data from the MIT-BIH arrhythmia database
(Mark et Moody, 1997). In particular, the considered
beats make reference to the following classes:
normal sinus rhythm (N), atrial premature beat (A),
ventricular premature beat (V), right bundle branch
block (RB), left bundle branch block (LB), and
paced beat (/). Similarly to (Inan et al., 2006), the
beats were selected from the recordings of 18
patients, which correspond to the following files:
100, 102, 104, 105, 106, 107, 118, 119, 200, 201,
203, 205, 208, 212, 213, 214, 215, and 217. For
feeding the classification process, we adopted in this
study the two following kinds of features: i) ECG
morphology features; and ii) three ECG temporal
features that are the QRS complex duration, the RR
interval (i.e., time span between two consecutive R
points representing the distance between the QRS
peaks of the present and previous beats), and the RR
interval averaged over the ten last beats (de Chazal
et Reilly, 2006). The total number of morphology
and temporal features is equal to 303 for each beat.
In order to train the classification process and to
assess its accuracy, we selected randomly from the
considered recordings 500 beats for the training set,
whereas 42185 beats were used as test set (thus, the
training set represents just 1.18% of the test set). The
detailed numbers of training and test beats are
reported for each class in Table 1. Classification
performance was evaluated in terms of three
accuracy measures, which are: 1) the overall
accuracy (OA); 2) the accuracy of each class; and 3)
the average accuracy (AA).
Due to the good performances generally
achieved by the nonlinear SVM classifier based on
the Gaussian kernel [6], we adopted this kernel in all
experiments. The parameters C and γ were varied in
the ranges [10
-3
, 200] and [10
-3
, 2], respectively. The
k value and the number of hidden nodes (h) of the
kNN and the RBF classifiers were tuned in the
intervals [1, 15] and [10, 60], respectively.
Concerning the PSO algorithm, we considered the
following standard parameters: swarm size S=40,
inertia weight w=0.4, acceleration constants c
1
and
c
2
equal to the unity, and maximum number of
iterations fixed to 40.
3.1 Experiment 1: Classification in the
Original Feature Space
In this experiment, we applied the SVM classifier
directly on the whole original hyperdimensional
feature space which is composed of 303 features.
During the training phase, the SVM parameters (i.e.,
C and γ) were selected according to a m-fold cross-
validation (CV) procedure. In all experiments
reported in this paper, we adopted a 5-fold CV. The
same procedure was adopted to find the best
parameters for the kNN and the RBF classifiers. The
best values obtained for the three investigated
classifiers are C=25, γ=0.5, k=3 and h=20. As
reported in Table 2, the OA and AA accuracies
achieved by the SVM classifier on the test set are
equal to 87.95% and 87.60%, respectively. These
results are much better than those achieved by the
RBF and the kNN classifiers. Indeed, the OA and
AA accuracies are equal to 82.78% and 82.34% for
the RBF classifier, and 78.21% and 79.34% for the
kNN classifier, respectively. This experiment
appears to confirm what observed in other
application fields, i.e., the superiority of SVM with
respect to traditional classifiers when dealing with
feature spaces of very high dimensionality.
3.2 Experiment 2: Classification based
on Feature Reduction
In this experiment, we trained the SVM classifier in
feature subspaces of various dimensionalities. The
desired number of features was varied from 10 to 50
with a step of 10, namely from small to high
dimensional feature subspaces. Feature reduction
was achieved by means of the traditional Principal
Component Analysis (PCA) algorithm. Figure 1-a
depicts the results obtained in terms of OA by the
three considered classifiers combined with the PCA
algorithm, namely the PCA-SVM, the PCA-RBF
and the PCA-kNN classifiers. In particular, it can be
seen that, for all feature subspace dimensionalities
except the lowest one (i.e., 10 features), the PCA-
SVM classifier maintains a clear superiority over the
two other classifiers. Its best accuracy was found by
using a feature subspace composed of the first 40
components. The corresponding OA and AA
accuracies are equal to 88.98% and 88%,
respectively. Comparing these results with those
obtained by the SVM classifier in the original
feature space (i.e., without feature reduction), a
slight increase of 1.03% and 0.4% in terms of OA
and AA, respectively, was achieved (see Table 2).
3.3 Experiment 3: Classification with
PSO-SVM
In this experiment, we applied the PSO-SVM
classifier on the available training beats. At
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
22
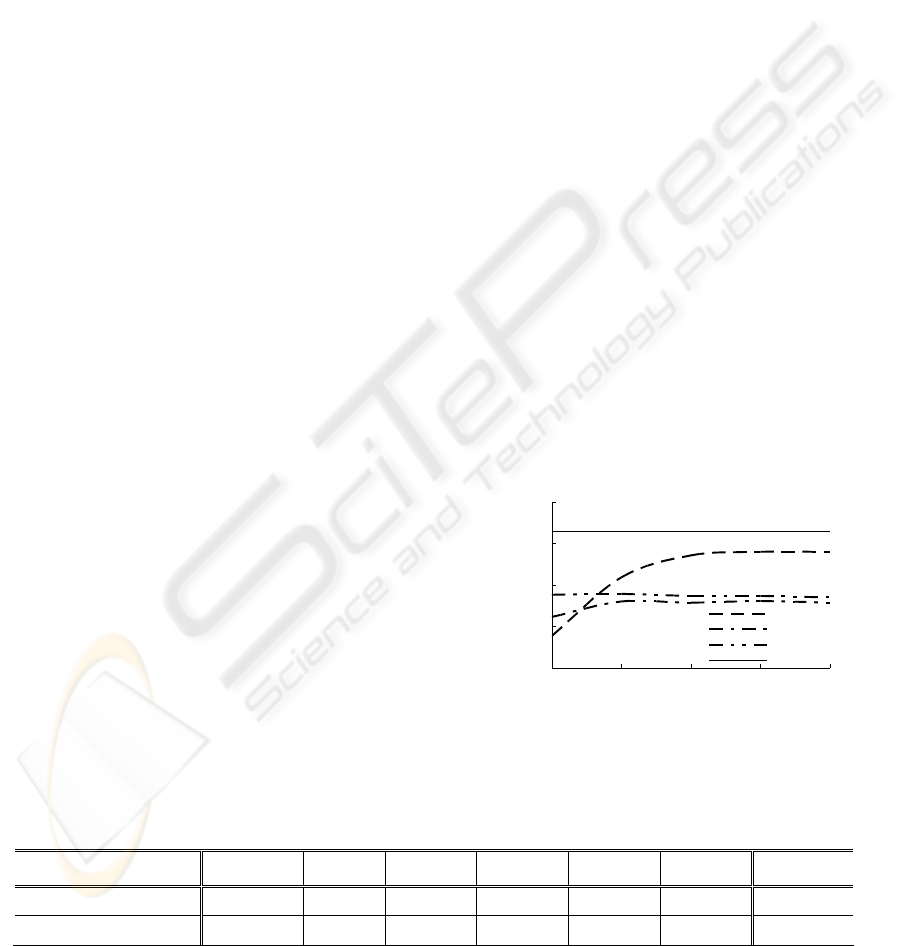
convergence of the optimization process, we
assessed the PSO-SVM classifier accuracy on the
test samples. The achieved overall and average
accuracies are equal to 91.44% and 91.19%
corresponding to substantial accuracy gains with
respect to what yielded either by the SVM classifier
applied on all available features (+3.49% and
+3.59%, respectively) or by the PCA-SVM classifier
(+2.46% and +3.19%, respectively) (see Table 2 and
Figure 1). Its worst class accuracy was obtained for
atrial premature beats (A) (88.16%) while that of the
SVM and the PCA-SVM classifiers corresponded to
paced beats (/) (73.43%) and ventricular premature
beats (V) (78.06%), respectively. This shows the
capability of the PSO-SVM classifier to reduce
significantly the gap between the worst and best
class accuracies (8.25% against 15.43% and 20.21%
for the PCA-SVM and the SVM classifiers,
respectively) while keeping the overall accuracy to a
high level.
4 CONCLUSIONS
The main novelty of this paper is to be found in the
proposed PSO-based approach that aims at
optimizing the performances of SVM classifiers in
terms of classification accuracy by detecting the best
subset of available features and by solving the tricky
model selection issue. Its completely automatic
nature renders it particularly useful and attractive.
The results confirm that the PSO-SVM classification
system boosts up significantly the generalization
capability achievable with the SVM classifier.
Finally, it is noteworthy that the general nature of
the proposed PSO-SVM system makes it applicable
not just to morphology and temporal features but to
other kinds of features such as those based on
wavelets and high-order statistics. Finally, other
optimization criteria could be considered as well
individually or jointly depending on the application
requirements.
REFERENCES
Bazi Y., Melgani F., (2006). Toward an Optimal SVM
Classification System for Hyperspectral Remote
Sensing Images. IEEE Trans. Geosci. Remote Sens.,
44, 3374-3385.
De Chazal F., Reilly R.B., (2006). A patient adapting heart
beat classifier using ECG morphology and heartbeat
interval features. IEEE Trans. Biomedical
Engineering, 43, 2535-2543.
El-Naqa I., Yongyi Y., Wernick M.N., Galatsanos N.P.,
Nishikawa R.M., (2002). A support vector machine
approach for detection of microcalcifications. IEEE
Trans. on Medical Imaging, 21, 1552-1563.
Inan O.T., Giovangrandi L., Kovacs J.T.A., (2006).
Robust neural network based classification of
premature ventricular contractions using wavelet
transform and timing interval features. IEEE Trans.
Biomedical Engineering, 53, 2507-2515.
Kennedy J., Eberhart R.C., (2001). Swarm Intelligence.
San Mateo, CA: Morgan Kaufmann.
Mark R., Moody G., (1997). MIT-BIH Arrhythmia
Database 1997 [Online]. Available:
http://ecg.mit.edu/dbinfo.html.
Melgani F., Bruzzone L., (2004). Classification of
hyperspectral remote sensing images with support
vector machine. IEEE Trans. on Geosci. Remote Sens.,
42, 1778-1790.
Pontil M., Verri A., (1998). Support vector machines for
3D object recognition”, IEEE Trans. on Pattern
Analysis and Machine Intelligence, 20, 637-646.
Vapnik V., (1998). Statistical Learning Theory. New
York: Wiley.
75,00
80,00
85,00
90,00
95,00
10 20 30 40 50
# Selected Features
OA [%]
PCA-SVM
PCA-RBF
PCA-KNN
PSO-SVM
Figure 1: Overall percentage accuracy (OA) versus
number of selected features (first principal components)
achieved on the test beats by the different classifiers.
Table 1: Numbers of training and test beats used in the experiments.
Class N A V RB / LB Total
Training beats 150 100 100 50 50 50 500
Test beats 24966 119 4239 3939 6971 1951 42185
EVOLUTIONARY COMPUTATION APPROACH TO ECG SIGNAL CLASSIFICATION
23
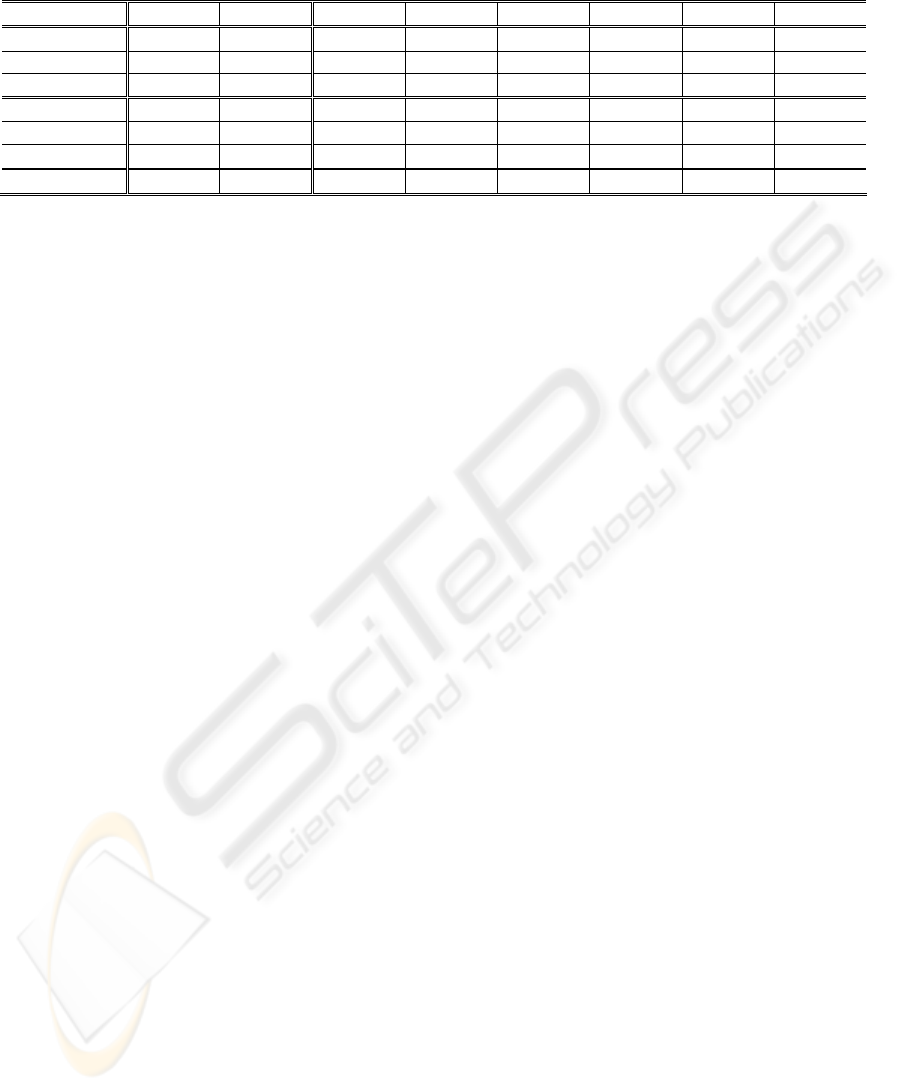
Table 2: Overall (OA), average (AA), and class percentage accuracies achieved on the test beats by the different
investigated classifiers.
Method OA AA N
A V RB / LB
SVM 87.95 87.60 90.05 83.19 92.12 93.15 73.43 93.64
RBF 82.78 82.34 85.14 78.99 90.39 86.74 66.53 86.26
kNN 78.21 79.34 76.50 66.38 71.99 93.27 75.92 92.00
PCA-SVM 88.98 88.00 89.36 83.19 78.06 93.50 90.60 93.28
PCA-RBF 83.04 82.11 85.86 80.67 87.85 83.87 68.85 85.54
PCA-kNN 83.91 82.02 85.62 69.74 79.05 93.04 73.89 90.77
PSO-SVM 91.44 91.19 91.12 88.16 93.70 93.70 92.01 96.41
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
24
