
MODERN CONCEPTS FOR
HIGH-PERFOMANCE SCIENTIFIC COMPUTING
Library Centric Application Design
Ren
´
e Heinzl, Philipp Schwaha and Siegfried Selberherr
Institute for Microelectronics, TU Wien, Gusshausstrasse 27-29, Vienna, Austria
Keywords:
Scientific computing, high performance computing, concept based programming, multi paradigms, partial
differential equations.
Abstract:
During the last decades various high-performance libraries were developed written in fairly low level lan-
guages, like FORTRAN, carefully specializing codes to achieve the best performance. However, the objective
to achieve reusable components has been regularly eluded by the software community ever since. The funda-
mental goal of our approach is to create a high-performance mathematical framework with reusable domain-
specific abstractions which are close to the mathematical notations to describe many problems in scientific
computing. Interoperability driven by strong theoretical derivations of mathematical concepts is another im-
portant goal of our approach.
1 INTRODUCTION
This work reviews common concepts for scientific
computing and introduces new ones for a timely ap-
proach to library centric application design.
Based on concepts for generic programming, e.g.
in C++, we have investigated and developed data
structures for scientific computing. The Boost Graph
Library (Siek et al., 2002) was one of the first generic
libraries, which introduced concept based program-
ming for a more complex data structure, a graph. The
actual implementation of the Boost Graph Library
(BGL) is for our work of secondary importance, how-
ever, we value the consistent interfaces for graph op-
erations. We have extended this type of concept based
programming and library development to the field of
scientific computing. To give a brief introduction we
use an example resulting from a self-adjoint partial
differential equation (PDE), namely the Poisson equa-
tion:
div(ε grad(Ψ)) = ρ
Several discretization schemes are available to
project this PDE into a finite space. We use the
method of finite volumes (FV (Selberherr, 1984)).The
resulting equations are given next, where A
ij
and d
ij
represents geometrical properties of the discretized
space, ρ the space charge, Ψ the potential, and ε the
permittivity of the medium.
∑
j
D
ij
A
ij
= ρ (1)
D
ij
=
Ψ
j
− Ψ
i
d
ij
ε
i
+ ε
j
2
(2)
An example of our domain specific notation is
given in the following code snippet and explained in
Section 4:
value =
(
sum<vertex_edge >
[
diff<edge_vertex >
[
Psi(_1)
] * A(_1)/d(_1) *
sum<edge_vertex >[eps(_1)]/2
] - rho(_1)
)(vertex);
Generic Poisson Equation
As can be seen, the actual notation does not de-
pend on any dimension or topological type of the cell
100
Heinzl R., Schwaha P. and Selberherr S. (2007).
MODERN CONCEPTS FOR HIGH-PERFOMANCE SCIENTIFIC COMPUTING - Library Centric Application Design.
In Proceedings of the Second International Conference on Software and Data Technologies - SE, pages 100-107
DOI: 10.5220/0001327401000107
Copyright
c
SciTePress

complex (mesh) and is therefore dimensionally and
topologically indepent. Only the relevant concepts,
in this case, the existence of edges incident to a ver-
tex and several quantity accessors, have to be met. In
other words, we have extended the concept program-
ming of the standard template library (STL) and the
generic programming of C++ to higher dimensional
data structures and automatic quantity access mecha-
nisms.
Compared to the reviewed related work given in
Section 2, our approach implements a domain spe-
cific embedded language. The related topological
concepts are given in Section 3, whereas Section 4
briefly overviews the used programming paradigms.
In Section 5 several application examples are pre-
sented. The first example introduces a problem of
a biological system with a simple PDE. The second
example shows a nonlinear system of coupled PDEs,
which makes use of the linearization framework intro-
duced in Section 4.1, where derivatives are calculated
automatically.
For a successful treatment of a domain specific
embedded notation several programming paradigms
are used. By object-oriented programming the ap-
propriate iterators are generated, hidden in this exam-
ple in the expression
vertex edge
and
edge vertex
.
Functional programming supplies the higher order
function expression between the
[
and
]
and the un-
named function object
1
. And finally the generic pro-
gramming paradigm (in C++ realized with parametric
polymorphism or templates) connects the various data
types of the iterators and quantity accessors.
A significant target of this work is the separa-
tion of data access and traversal by means of the
mathematical concept of fiber bundles (Butler and
Bryson, 1992). The related formal introduction en-
ables a clean separation of the internal combinatorial
properties of data structures and the mechanisms of
data access. A high degree of interoperability can be
achieved with this formal interface. Due to space con-
straints the performance analysis is omitted and we
refer to a recent work (Heinzl et al., 2006a) where the
overall high performance is presented in more detail.
2 RELATED WORK
In the following several related works are presented.
All of these software libraries are a great achievement
in the various fields of scientific computing.
The FEniCS project (Logg et al., 2003), which is
a unified framework for several tasks in the area of
scientific computing, is a great step towards generic
modules for scientific computing.
Femster (Castillo et al., 2005) is a class library
for finite element (FE) calculations. This means that
users must provide their own code for assembling
global FE matrices. In other words, Femster imple-
ments a general finite element API.
The Template Numerical Toolkit (Pozo, 1997)
is a collection of interfaces and reference implemen-
tations of numerical objects (matrices) in C++. The
toolkit defines interfaces for basic data structures,
such as multidimensional arrays and sparse matrices,
commonly used in numerical applications.
The Boost Graph Library is a generic interface
which enables access to a graph’s structure, but hides
the details of the actual implementation. All libraries
which implement this type of interface are interoper-
able with the BGL generic algorithms. This approach
was one of the first in the field of non-trivial data
structures with respect to interoperability. The prop-
erty map concept (Siek et al., 2002) was introduced
and heavily used.
The Grid Algorithms Library, GrAL (Berti,
2000) was one of the first contributions to the uni-
fication of data structures of arbitrary dimension for
the field of scientific computing. A common interface
for grids with a dimensionally and topologically inde-
pendent way of access and traversal was designed.
Our approach, the Generic Scientific Simulation
Environment, GSSE (Heinzl et al., 2006b) deals with
various modules for different discretization schemes
such as finite elements and finite differences. In
comparison, our approach focuses more on providing
building blocks for scientific computing, especially an
embedded domain language to express mathematical
dependencies directly, not only for finite elements.
To achieve interoperability between different li-
brary approaches we use concepts of the fiber bun-
dle theory to separate the base space and fiber space
properties. With this separation we can use several
other libraries (see Section 3) for different tasks. The
theory of fiber bundles separates the data structural
components from data access (fibers). We have de-
veloped a consistent data structure interface for all
different types of data structures and several other li-
braries employing the theory of CW-complexes and
poset theory. Based on this interface specification we
can use several libraries, such as STL, BGL, GrAL,
and accomplish high interoperability and code reuse.
3 CONCEPTS
Our approach extends the concept based program-
ming of the STL to arbitrary dimensions similar to
GrAL. The main difference to GrAL is the introduc-
MODERN CONCEPTS FOR HIGH-PERFOMANCE SCIENTIFIC COMPUTING - Library Centric Application Design
101
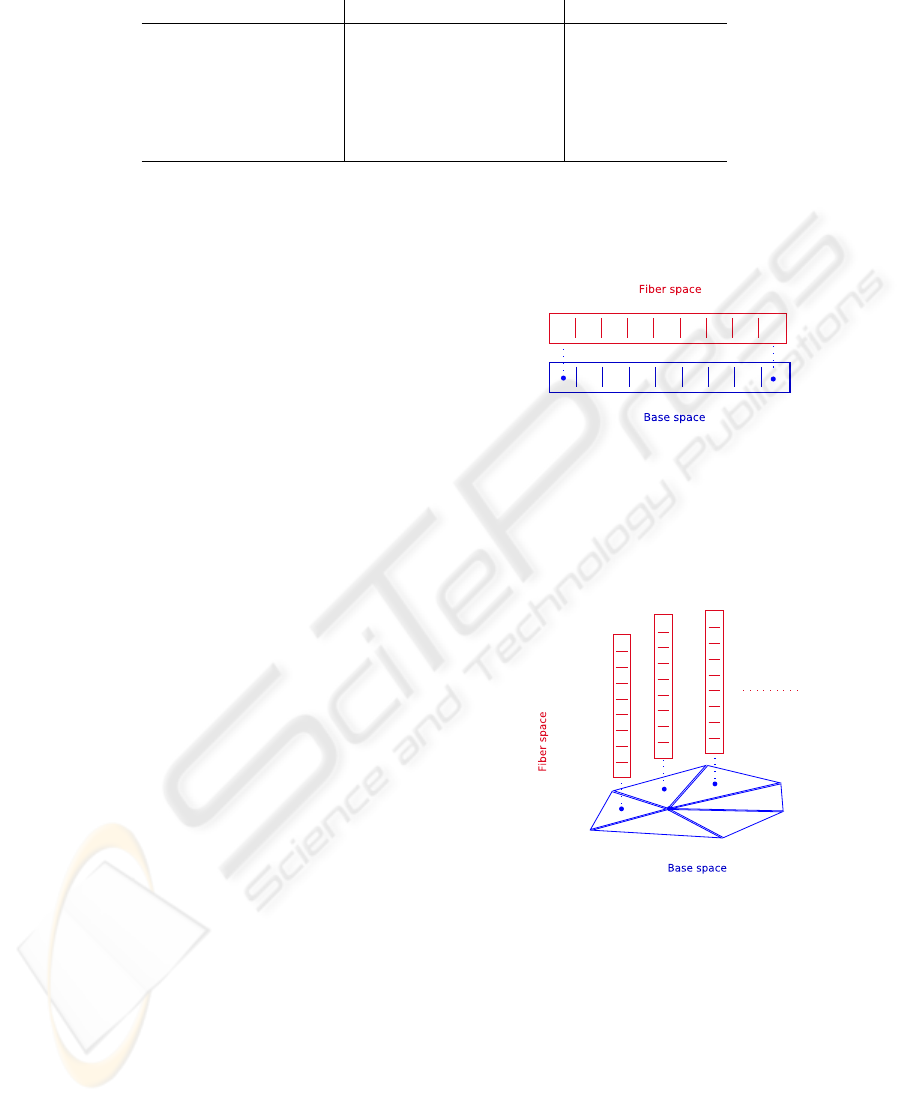
Table 1: Comparison of the cursor/property map and the fiber bundle concept.
cursor and property map fiber bundles
isomorphic base space no yes
traversal possibilities
STL iteration cell complex
traversal base space
yes yes
traversal fiber space
no yes
data access
single data topological space
fiber space slices
no yes
tion of the concept of fiber bundles, which separates
the base mechanism of application design into base
and fiber space properties. The base space is modeled
by a CW-complex and algebraic topology, whereas
the fiber space is modeled by a generic data accessor
mechanism, similar to the cursor and property map
concept (Abrahams et al., 2003).
3.1 Theory of Fiber Bundles
We introduce concepts of fiber bundles as a descrip-
tion for data structures of various dimensions and
topological properties.
• Base space: topology and partially ordered sets
• Fiber space: matrix and tensor handling
Based on these examples, we introduce a com-
mon theory for the separation of the topological struc-
ture and the attached data. The original contribu-
tion of this theory was given in Butler’s vector bun-
dle model (Butler and Bryson, 1992), which we com-
pactly review here:
Definition 1 (Fiber Bundle) Let E,B be topological
spaces and f : E → B a continuous map. Then
(E,B, f) is called a fiber bundle, if there exists a
space F such that the union of the inverse images of
the projection map f (the fibers) of a neighborhood
U
b
⊂ B of each point b ∈ B are homeomorphic to
U
b
× F, whereby this homeomorphism has to be such
that the projection pr
1
of U
b
× F (that maps each el-
ement of this product space to the element of the first
space) yields U
b
again.
E is called the total space, B is called the base space,
and F is called the fiber space. This definition re-
quires that a total space E can locally be written as
the product of a base space B and a fiber space F.
The decomposition of the base space is mod-
eled by an identification of data structures by a CW-
complex. (Benger, 2004; Heinzl et al., 2006c). As an
example Figure 2 depicts an array data structure based
on the concept of a fiber bundle. We have a simple
fiber space attached to each cell (marked with a dot in
the figure), which conserves the neighborhood of our
base space and carries the data of our array.
Figure 2: A fiber bundle with a fiber space over a 0-cell
complex. A simple array is an example of this type of fiber
space.
The next figure depicts a fiber bundle with a 2-
cell complex as base space. For the base space of
Figure 3: A fiber space over a 2-simplex cell complex base
space. An example of this type of fiber space is a triangle
mesh with an array over each triangle.
lower dimensional data structures, such as an array or
single linked list, the only relevant information is the
number of elements determined by the index space.
Therefore most of the data structures do not separate
these two spaces. For backward compatibility with
common data structures the concept of an index space
depth is used (Heinzl et al., 2006c).
The advantages of this approach are similar to
those of the cursor and property map (Abrahams et al.,
2003), but they differ in several details. The similar-
ICSOFT 2007 - International Conference on Software and Data Technologies
102
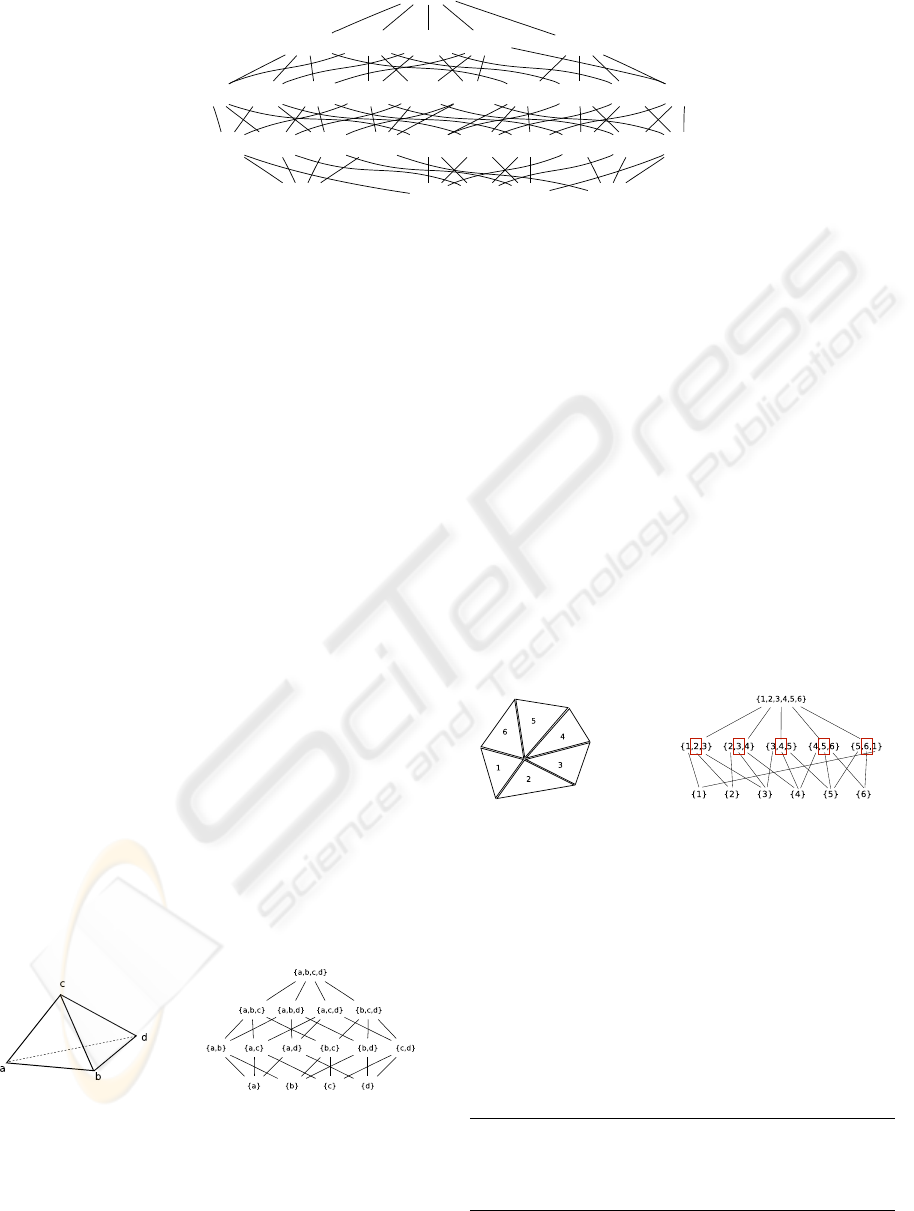
{0,1,2,3,4}
{0,1,2,3}
{0,1,2,4} {0,1,3,4}{0,2,3,4} {1,2,3,4}
{0,1,2} {0,1,3}{0,2,3} {1,2,3}{0,1,4}{0,2,4} {1,2,4}{0,3,4} {1,3,4}{2,3,4}
{0,1}{0,2} {1,2}{0,3} {1,3}{2,3}{0,4} {1,4}{2,4}
{0} {1}{2}
{3,4}
{3} {4}
Figure 1: Cell topology of a simplex cell in four dimensions.
ity is that both properties can be implemented inde-
pendently. However, the fiber bundle approach equips
the fiber space with more structure, e.g., storing more
than one value corresponding to the traversal position
as well as preservation of neighborhoods. This feature
is especially useful in the area of scientific computing,
where different data sets have to be managed, e.g.,
multiple scalar or vector values on vertices, faces, or
cells. Another important property of the fiber bundle
approach is that an equal (isomorphic) base space can
be exchanged with another cell complex of the same
dimension. Table 1 summarizes the common features
and the differences.
As can be seen the concept of the cursor and prop-
erty map can be extended to the fiber bundle approach
with additional properties and mechanisms.
3.2 Topological Interface
We briefly introduce parts of the interface specifica-
tion for data structures and their corresponding itera-
tion mechanism based on algebraic topology. A full
reference can be found in (Heinzl et al., 2006c). With
the concept of partial ordered sets and a Hasse dia-
gram we can order and depict the structure of a cell.
As an example the topological structure of a three-
dimensional simplex is given in Figure 4. Inter-
Figure 4: Cell topology of a 3-simplex cell.
dimensional objects such as edges and facets and their
relations within the cell can thereby identified. The
complete traversal of all different objects is deter-
mined by this structure. We can derive the vertex on
edge, vertex on cell, as well as edge on cell traver-
sal up to the dimension of the cell in this way. Based
on our topological specification arbitrary dimensional
cells can be easily used and traversed in the same way
as all other cell types, e.g., a 4-dimensional simplex
shown in Figure 1.
Next to the cell topology a separate complex
topology is derived to enable an efficient implemen-
tation of these two concepts. A significant amount of
code can be reduced with this separation. Figure 5 de-
picts the complex topology of a 2-simplex cell com-
plex where the bottom sets on the right-hand sides are
now the cells. The rectangle in the figure marks the
relevant cell number. The topology of the cell com-
Figure 5: Complex topology of a simplex cell complex.
plex is only available locally because of the fact that
subsets can have an arbitrary number of elements. In
other words, there can be an arbitrary number of trian-
gles attached to the innermost vertex. Our final classi-
fication scheme uses the term
local
to represent this
fact.
A formal concise definition of data structures can
therewith be derived and is presented in Figure 2. The
complex topology uses the number of elements of the
corresponding subsets.
complex_t <cells_t ,global > cx; //{1}
complex_t <cells_t ,local<2>> cx; //{2}
complex_t <cells_t ,local<3>> cx; //{3}
complex_t <cells_t ,local<4>> cx; //{4}
STL Data Structure Definitions
MODERN CONCEPTS FOR HIGH-PERFOMANCE SCIENTIFIC COMPUTING - Library Centric Application Design
103
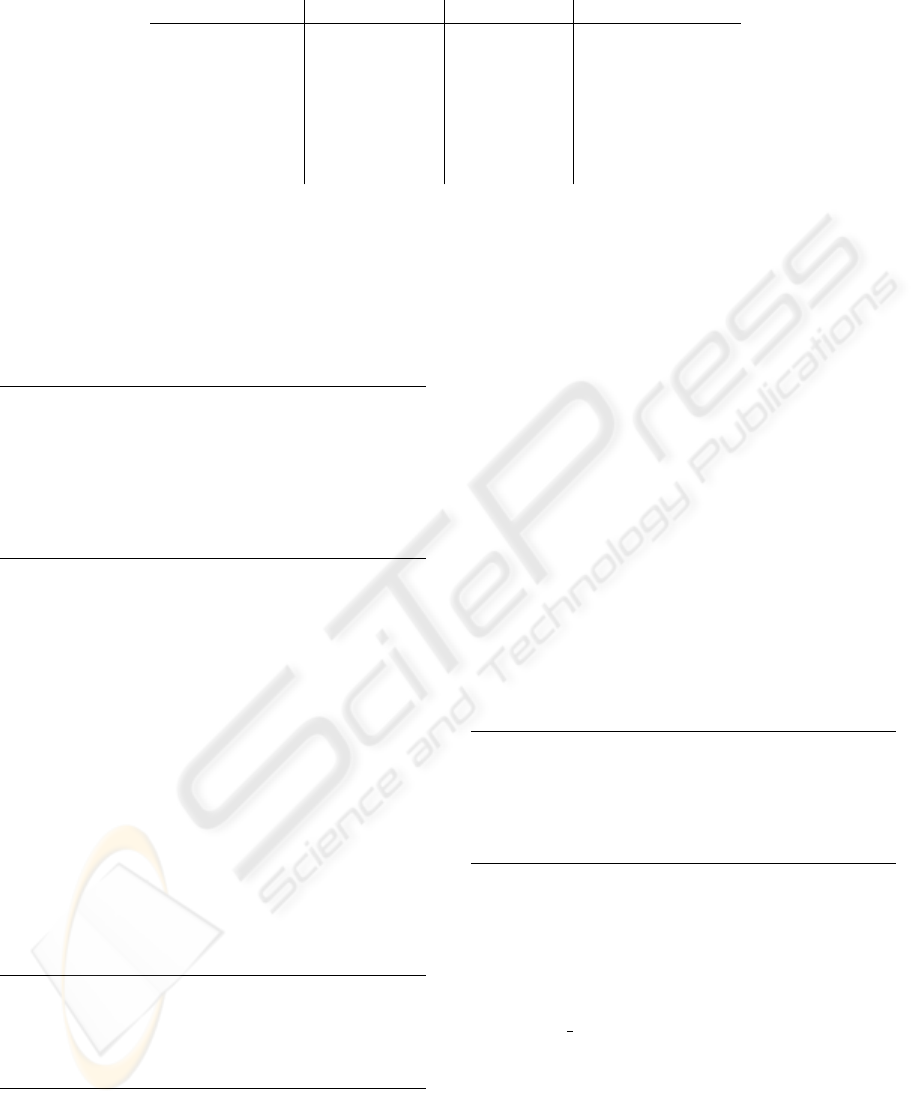
Table 2: Classification scheme based on the dimension of cells, the cell topology, and the complex topology.
data structure
cell dimension cell topology complex topology
array/vector 0 simplex global
SLL/stream
0 simplex local(2)
DLL/binary tree
0 simplex local(3)
arbitrary tree
0 simplex local(4)
graph
1 simplex local
grid
2,3,4,.. cuboid global
mesh
2,3,4,.. simplex local
Here {
1
} describes an array, {
2
} a stream or a sin-
gle linked list, {
3
} a doubly linked list or a binary
tree, and finally {
4
} an arbitrary tree. To demonstrate
the equivalence of the STL data structures and our ap-
proach we present a simple code snippet (the typedefs
are omitted due to space constraints):
cell_t <0, simplex> cells_t;
complex_t <cells_t , global > complex_t;
long
data_t;
container_t <complex_t , data_t >
container;
// is equivalent to
std::vector <data_t> container;
Equivalence of Data Structures
The separation of the topological structure and the
data specification can be seen clearly here.
3.3 Data Access
In the following code snippet a simple example of the
generic use of a data accessor similar to the property
map concept is given, where a scalar value is assigned
to each vertex. The data accessor implementation
also takes care of accessing data sets with different
data locality, e.g., data on vertices, edges, facets, or
cells. The data accessor is extracted from the con-
tainer to enable a functional access mechanism with a
key value which can be modeled by arbitrary compa-
rable data types.
da_type da(container , key_d);
for_each(container.vertex_begin(),
container.vertex_end(),
da = 1.0 );
Data Accessor
Several programming paradigms are used in this
example which are presented in detail in the next sec-
tion, especially the functional programming, given in
this example with the
da = 1.0
.
4 PROGRAMMING PARADIGMS
Various areas of scientific computing encourage dif-
ferent programming techniques, even demands for
several programming paradigms can be observed:
• Object-oriented programming: the close interac-
tion of content and functions is one of the most
important advantages of the object-oriented pro-
gramming
• Functional programming: offers a clear notation,
is side-effect free and inherent parallel
• Generic programming: can be seen as the glue
between object-oriented and functional program-
ming
Our implementation language of choice is C++
due to the fact, that it is one of the few programming
languages where high performance can be achieved
with various paradigms. To give an example of this
multi-paradigm approach, a simple C++ source snip-
pet is given next.
std::for_each(v.begin(),v.end(),
if_(arg1 > 5)
[
std::cout << arg1 << std::cout
]
);
Multiple Paradigms
The object-oriented programming paradigm is used to
create the iterator capabilities of the container struc-
tures of the STL. This paradigm is not used anywhere
else in our approach. Functional programming is used
to create function objects which are passed into the
generic
for each
algorithm. In this example the no-
tation of the Boost Phoenix 2 (Pho, 2006) library is
used to create a functional object context, marked by
the
[
and
]
. The generic paradigm uses the template
mechanism of C++ to bind these two paradigms to-
gether efficiently. A more complex example is given
in the following expression. Here a cell complex of
arbitrary dimension is used and the vertex to vertex
iteration is expressed.
ICSOFT 2007 - International Conference on Software and Data Technologies
104
gsse::for_each((*segit).vertex_begin(),
(*segit).vertex_end(),
result=gsse::add<vertex_vertex >
(
_1+_2
)[quan]
);
Complex Functor
The same paradigms as in the example before can
be seen, but in this case, a complex topological traver-
sal is used instead of simple container traversal. The
topological properties of the GSSE are demonstrated
twofold: on the one hand side, the topological concept
programming allows the implementation of a dimen-
sionally independend algorithm. On the other hand
side, different data structures of library approaches
can be used, which fullfill the basic requirements of
the required topological concept. In this example all
data structures or libraries which implement means
of vertex to vertex traversal can be used. The func-
tional expression is also more complex and based on
the algebraic property of the identity element for the
gsse::add
operation as the initial value of the opera-
tion, in this case
0
. This GSSE algorithm sums up the
potential values of all adjacent vertices to a vertex.
The data accessor
quan
handles the storage mecha-
nism for the value attached to a vertex. Here the inter-
action of programming paradigms related to the base
space and fiber space can be seen clearly. The base
space traversal is built with the generic programming
paradigm, whereas the fiber space operation is imple-
mented by means of functional programming.
A lot of difficulties with conventional program-
ming can be circumvented by this approach. Func-
tional programming enables great extensibility due
to the modular nature of function objects. Generic
programming and the corresponding template mech-
anisms of C++ offer an overall high performance. In
addition arbitrary data structures of arbitrary dimen-
sions can be used. The only requirement is that the
data structure models the required concept, in this
case a vertex to vertex information.
4.1 Automatic Linearization
We introduce a calculation framework where deriva-
tives are implicitly available and do not have to be
specified explicitly. This enables the specification of
nonlinear differential equations in a convenient way.
The elements of the framework are truncated Taylor
series of the following form f
0
+
∑
i
c
i
· ∆x
i
. To use
a quantity x
i
within a formula we have to specify its
value f
0
and the linear dependence c
i
= 1 on the vec-
tor x of quantities. This step, however, can be per-
formed implicitly by the computer. This non-trivial
and highly complex scenario yields itself exception-
ally well to the application of the functional program-
ming paradigm. In general, all discretization schemes
which use line-wise assembly based on finite differ-
ences as well as finite volumes can be handled with
the described formalism.
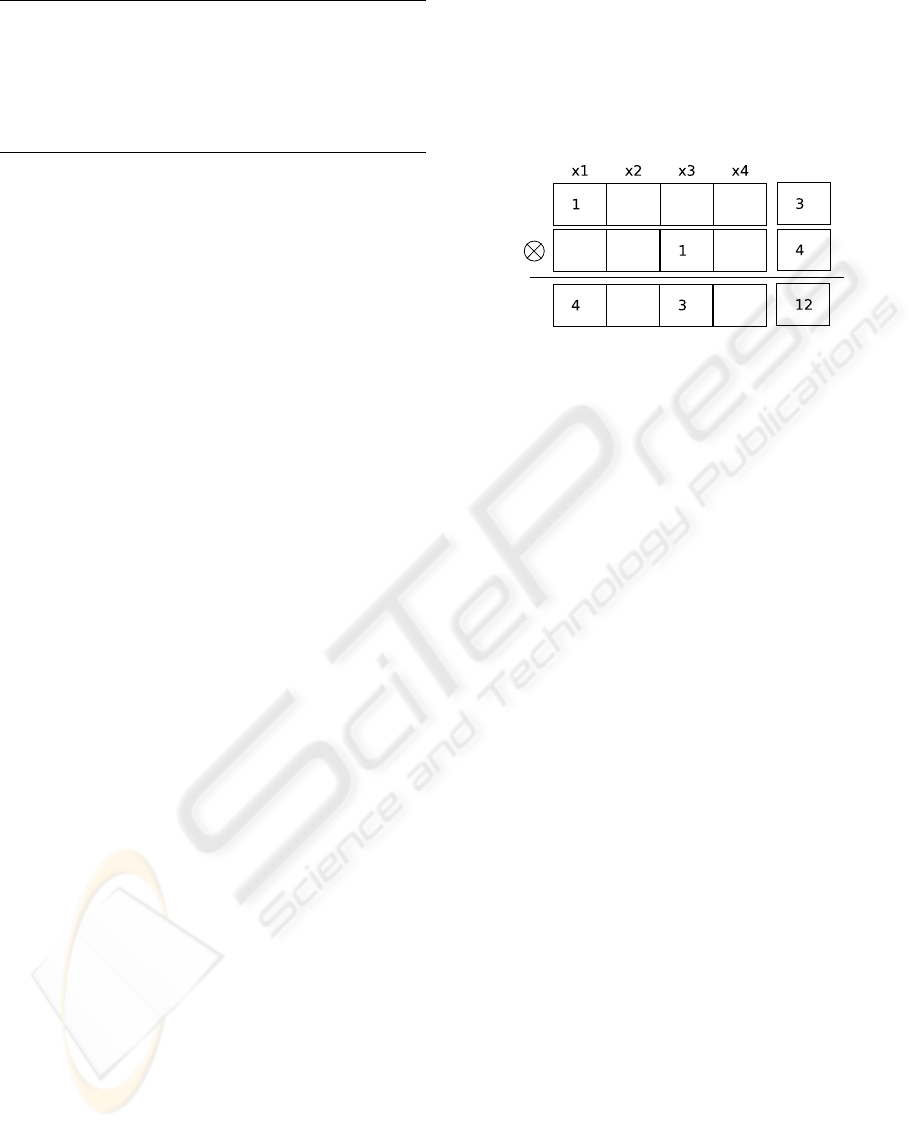
Figure 6: The multiplication of two Taylor series.
Basic operations on Taylor series can handle trun-
cated polynomial expansions. In the following we
specify our nonlinear functionals using linearized
functions in upper case letters. All necessary numer-
ical operations on these data structures can be per-
formed in a straight forward manner.
F = f
0
+
∑
i
c
i
· ∆x
i
, G = g
0
+
∑
i
d
i
· ∆x
i
(3)
F ⊕ G = ( f
0
+ g
0
) +
∑
i
(c
i
+ d
i
) · ∆x
i
(4)
F ⊗ G = ( f
0
· g
0
) +
∑
i
(g
0
· d
i
+ f
0
· c
i
) · ∆x
i
(5)
Having implemented these schemes we are able
to derive all required functions on these mathematical
structures. This means that we have a consistent
framework for formulas in the following sense: if A is
the linearized version of function
A at x
0
, we obtain
∂A/∂
x
i
= ∂
A /∂
x
i
|
x
0
around the point of linearization.
Figure 6 shows the multiplication of two truncated
expansions F = 3+ ∆x
1
, and G = 3+ ∆x
3
. As a re-
sult we obtain F ⊗ G = 12 + 4∆x
1
+ 3∆x
3
. By im-
plementing only the linear (or higher order polyno-
mial) functional dependence of equations on variables
around x we reduce the external specification effort
to a minimum. Thus, it is possible to ease the spec-
ification with the functional programming approach,
while also providing the functional dependence of
formulas.
5 APPLICATION DESIGN
In the following we briefly review a few applications
based on the introduced concepts with their respective
paradigms.
MODERN CONCEPTS FOR HIGH-PERFOMANCE SCIENTIFIC COMPUTING - Library Centric Application Design
105
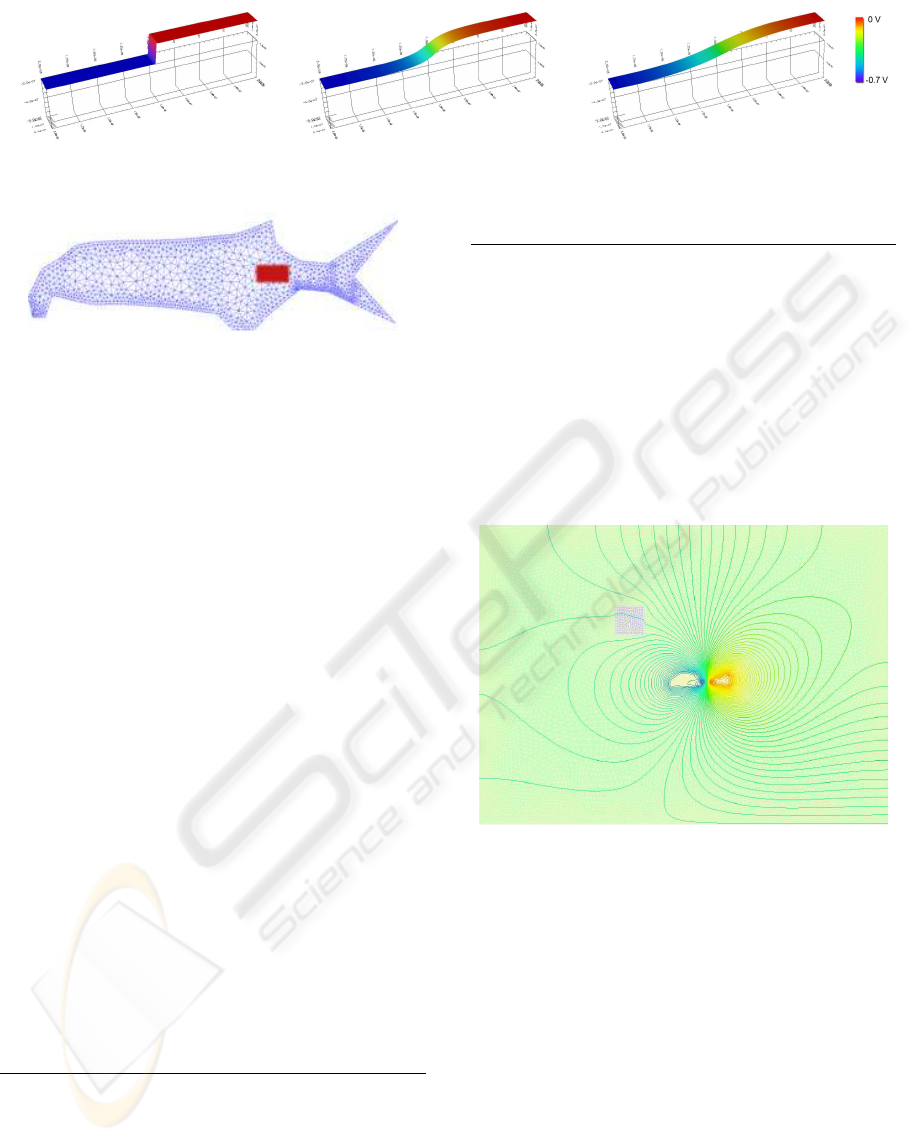
Figure 8: Potential in a pn diode during different stages of the Newton iteration. From initial (left) to the final result(right).
Figure 7: Discretized domain of a fish with a red marked
electrically active organ.
5.1 Biological System
Electric phenomena are common in biological organ-
isms such as the discharges within the nervous sys-
tem, but usually remain within a small scale. In some
organisms, however, the electric phenomena take a
more prominent role. Some fish species, such as
Gnathonemus petersii from the family of Mormyridae
(Westheide and Rieger, 2003), use them for detection
of their prey. The up to 30 cm long fish actively gener-
ates electric pulses with an organ located near its tail
fin (also marked in Figure 7). More information can
be found in (Schwaha et al., 2007).
For this case we derive the equation system based
on a quasi-electro-statical system directly from the
corresponding Maxwell equations. The charge sep-
aration of the electrically active organ which is ac-
tively taking place within parts of the simulation do-
main also has to be taken into account. We use the
conservation law of charge and the divergence theo-
rem (Gauss’s law) and finally get:
∂
t
[div(ε grad(Ψ))] + div(γ grad(Ψ)) = P (6)
Equation 6 is discretized using the finite volume dis-
cretization scheme. The high semantic level of the
specification is illustrated by the following snippet of
code:
equation=sum<vertex_edge >(_v)
[
Orient(_v,_1) *
sum<edge_vertex >(_e)
[
lineqn(pot(_1), psi(_1))*Orient(_e,_1)
]
*(area(_1) / dist(_1))
*(gamma(_1)*deltat + eps(_1))
]+vol(_1)*((P(_1)*deltat)+rho(_1)))
This source snippets presents most of the applica-
tion code which has to be developed. In addition, only
a simple preprocessing step which creates the neces-
sary quantity accessors is required.
The simulation domain is divided into several
parts including the fish itself, its skin, that serves as
insulation, the water the fish lives in, and an object,
that represents either an inanimate object or prey. The
parameters of each part can be adjusted separately. A
result of the simulation is depicted in the following
figure:
Figure 9: Result of a simulation with a complete domain,
the fish, and a ideally conductor as response object.
5.2 Drift-Diffusion Equation
Semiconductor devices have become an ubiquitous
commodity and people expect a constant increase of
device performance at higher integration densities and
falling prices.
To demonstrate the importance of a method for
device simulation that is both easy and efficient we
review the drift diffusion model that can be derived
from Boltzmann’s equation for electron transport by
applying the method of moments (Selberherr, 1984).
Note that this problem is a nonlinear coupled sys-
tem of partial differential equations where our lin-
earization framework is used. This results in current
ICSOFT 2007 - International Conference on Software and Data Technologies
106

relations as shown in Equation 7. These equations
are solved self consistently with Poisson’s equation,
given in Equation 8.
J
n
= qnµ
n
grad Ψ+ qD
n
grad n (7)
div(grad(ε Ψ)) = −ρ (8)
The following source code results from an application
of the finite volume discretization scheme:
linearequ_t equ_pot, equ_n;
equ_pot = (sum<vertex_edge >
[
diff<edge_vertex >[pot_quan]
] + ( n_quan - p_quan + nA - nD ) *
vol * q / (eps0 * epsr)
)(vertex);
equ_n = (sum<vertex_edge >
[
diff<edge_vertex >
( -n_quan*Bern(
diff<edge_vertex >[pot_quan /U_th]
),
-n_quan*Bern(
diff<edge_vertex >[-pot_quan/U_th]
)
)* (q * mu_h * U_th)
])(vertex);
Drift-Diffusion Equation
To briefly present a simulation result we provide
Figure 8 which shows the potential in a pn diode
at different stages of a nonlinear solving procedure.
The leftmost figure shows the initial solution, while
the rightmost depicts the final solution. The center
image shows an intermediate result that has not yet
fully converged. The visualization of the calculation
is available in real time, making it possible to ob-
serve the evolution of the solution, which is realized
by OpenDX (DX, 1993).
REFERENCES
DX (1993). IBM Visualization Data Explorer. IBM Corpo-
ration, Yorktown Heights, NY, USA, third edition.
Phoenix2 (2006). Boost Phoenix 2. Boost C++ Libraries.
http://spirit.sourceforge.net/.
Abrahams, D., Siek, J., and Witt, T. (2003). New Iterator
Concepts. Technical Report N1477 03-0060, ISO/IEC
JTC 1, Information Technology, Subcommittee SC
22, Programming Language C++.
Benger, W. (2004). Visualization of General Relativistic
Tensor Fields via a Fiber Bundle Data Model. Doc-
toral thesis, Freie Universit
¨
at Berlin.
Berti, G. (2000). Generic Software Components for Scien-
tific Computing. Doctoral thesis, Technische Univer-
sit
¨
at Cottbus.
Butler, D. M. and Bryson, S. (1992). Vector Bundle Classes
From Powerful Tool for Scientific Visualization. Com-
puters in Physics, 6:576–584.
Castillo, P., Rieben, R., and White, D. (2005). FEMSTER:
An Object-Oriented Class Library of High-Order Dis-
crete Differential Forms. ACM Trans. Math. Softw.,
31(4):425–457.
Heinzl, R., Schwaha, P., Spevak, M., and Grasser, T.
(2006a). Performance Aspects of a DSEL for Scien-
tific Computing with C++. In Proc. of the POOSC
Conf., pages 37–41, Nantes, France.
Heinzl, R., Spevak, M., Schwaha, P., and Grasser, T.
(2006b). A High Performance Generic Scientific Sim-
ulation Environment. In Proc. of the PARA Conf.,
page 61, Umea, Sweden.
Heinzl, R., Spevak, M., Schwaha, P., and Selberherr, S.
(2006c). A Generic Topology Library. In Library
Centric Sofware Design, OOPSLA, pages 85–93, Port-
land, OR, USA.
Logg, A., Dupont, T., Hoffman, J., Johnson, C., Kirby,
R. C., Larson, M. G., and Scott, L. R. (2003). The
FEniCS Project. Technical Report 2003-21, Chalmers
Finite Element Center.
Pozo, R. (1997). Template Numerical Toolkit for Linear
Algebra: High Performance Programming with C++
and the Standard Template Library. 11(3):251–263.
Schwaha, P., Heinzl, R., Mach, G., Pogoreutz, C., Fister,
S., and Selberherr, S. (2007). A High Performance
Webapplication for an Electro-Biological Problem. In
Proc. of the 21th ECMS 2007, Prague, Czech Rep.
Selberherr, S. (1984). Analysis and Simulation of Semicon-
ductor Devices. Springer, Wien–New York.
Siek, J., Lee, L.-Q., and Lumsdaine, A. (2002). The Boost
Graph Library: User Guide and Reference Manual.
Addison-Wesley.
Westheide, W. and Rieger, R. (2003). Spezielle Zoologie.
Teil 2: Wirbel- oder Sch
¨
adeltiere. Elsevier.
MODERN CONCEPTS FOR HIGH-PERFOMANCE SCIENTIFIC COMPUTING - Library Centric Application Design
107
