
Improved Singular Value Decomposition for Supervised
Learning in a High Dimensional Dataset
Ricco Rakotomalala
1
and Faouzi Mhamdi
2
1
ERIC Laboratory - University of Lyon 2
Bron, France
2
URPAH - University of Tunis
Tunis, Tunisie
Abstract. Singular Value Decomposition (SVD) is a useful technique for di-
mensionality reduction with a controlled loss of information. This paper makes
the very simple but worth-while observation that many attributes that contain no
information about the class label, may thus be selected erroneously for a super-
vised learning task. We propose to first use a very tolerant filter to select on a
univariate basis which attributes to include in the subsequent SVD. The features,
“the latent variables”, extracted from relevant descriptors allow to build a better
classifier with a significant improvement of the generalization error rate and less
cpu time. We show the efficiency of this combination of feature selection and
construction approaches on a protein classification context.
1 Introduction
Data preprocessing is a crucial step when we have to analyze an unstructured dataset.
Indeed, it is not possible to handle directly the native description of data to run a ma-
chine learning algorithm when we treat images, text, or in our case, when we want to
predict proteins families from their primary structure. The learning process is thus pre-
ceded by two data preprocessing operations: extract descriptors from the native format
of data in order to build an attribute value table; build features from these descriptors in
order to produce an efficient classifier
3
.
The direct use of all descriptors extracted from the unstructured representation as
features for the learning algorithm is in general not a good strategy. Their number is
very high, which induces drawbacks: the computing time is very high and the quality of
the learning classifier is often poor because we have a sparse dataset, and it is difficult
to estimate in a reliable way the probability distribution (”The curse of Dimensionality
Problem”). In a protein discrimination process from their primary structures [1], the
native description of a protein is a succession of characters representing amino acids.
It is not possible to run directly a learning algorithm. We then generated a Boolean
3
In this paper, we call ”descriptors” the attributes which are extracted from the native data
format, i.e., n-grams in our context; we call ”features” the attributes which are presented to
the supervised learning algorithm.
Rakotomalala R. and Mhamdi F. (2006).
Improved Singular Value Decomposition for Supervised Learning in a High Dimensional Dataset.
In 6th International Workshop on Pattern Recognition in Information Systems, pages 38-47
DOI: 10.5220/0002472600380047
Copyright
c
SciTePress

attribute-value table by checking the presence or absence of 3-grams (a sequence of
3 consecutive characters) for each protein. Because there are 20 kinds of characters
(amino acids), we can produce 8000 descriptors for 100 examples, the quality of the
classifier on all descriptors is often bad.
To solve these disadvantages, we are interested in the creation of intermediate fea-
tures from the descriptors. The goal is to produce a new representation space which
preserves the properties of the initial space, in particular by preserving the proximity
between the examples. These new features, which will be provided to the learning al-
gorithm, must have the following qualities: they must represent a good summary of the
original data; they must be easy to interpret so that we can understand the influence of
each descriptors; they must be relevant for a supervised learning task; a small number
of them must be sufficient to learn classifier efficiently. The singular value decomposi-
tion (SVD) seems to answer in an adequate way these specifications. Indeed it aims to
transform raw data to a new co-ordinate system, where the axes of the new space repre-
sent “factors” or “latent variables” which reveal the underlying structure of the dataset.
This approach, very popular in high dimensional data processing, presents nevertheless
a drawback in the context of supervised learning: a lot of initial descriptors are irrele-
vant for the supervised learning task. To take them into account in the construction of
features (factors) considerably reduced the relevance of these features.
In this paper, we propose to insert a phase of descriptor selection before building
the latent variables with the singular value decomposition. This phase of selection must
only take account of the relevance of the descriptors and not of their redundancy, it
must be rather permissive so that information necessary to discriminate is preserved.
Only the selected descriptors will then be presented to the SVD, thus making it possi-
ble to produce an effective reduced space of representation for discrimination. In our
protein discrimination context, the results show that it is sufficient to keep 5 factors.
Another advantage, although that was not our first goal in this work, is that the reduc-
tion of the number of descriptors presented to the SVD algorithm allows one to reduce
dramatically the computing time.
Section 2 introduces the SVD process and our improvement in the context of su-
pervised learning in high dimensional dataset. The protein discrimination problem and
results of experiments are presented in Section 3. Section 4 describes some further ex-
periments which allows us to better evaluate the behavior of our approach. We conclude
in Section 5.
2 The Singular Value Decomposition for Supervised Learning
2.1 The Singular Value Decomposition Process
SVD produces a new representation space of the observations starting from the ini-
tial descriptors by preserving the proximity between the examples. These new features
known as ”factors” or ”latent variables” have several very advantageous properties: (a)
their interpretation very often allows to detect patterns in the initial space; (b) a very
reduced number of factors allows to restore information contained in the data; (c) the
new features form an orthogonal basis, learning algorithms such as linear discriminant
39

analysis work well [2]. This process is often used in microarray data analysis [3] or text
retrieval [4], fields where the initial number of descriptors is very high and where the
dimensionality reduction is crucial before data analysis.
There are numerous theoretical presentations of the SVD. Roughly speaking, we
produce from an initial description space ℵ = {X
1
, . . . , X
J
} of J descriptors (and n
examples), a new space of J features Ψ = {F
1
, . . . , F
J
} with the following constraints:
Ψ is an orthogonal basis; the factor F
1
is built from a projection vector P
1
(kP
1
k = 1)
so as to maximize the variance of F
1
, v
1
= Var (F
1
); the second factor F
2
is built from
a projection vector P
2
(kP
2
k = 1) so as to maximize the variance v
2
= Var (F
2
), and
F
2
must be independent (perpendicular) to F
1
, etc. In the two spaces, the proximity
between two individuals is preserved, and more interesting, in the subspace p (p <
J) of Ψ, the distance between two examples is roughly respected, the quality of the
approximation can be measured using the sum of variance of p first selected factors
(S
p
=
P
p
j=1
v
j
).
There is a mathematical relation between SVD and PCA (Principal Component
Analysis) when the descriptors are standardized. If ℵ
′
is the transpose of ℵ, the square
matrix (ℵ
′
ℵ) is a correlation matrix: v
1
is its first eigenvalue and P
1
is the associated
eigenvector. Thus, the sum of variance of the first p selected factors is the proportion of
explained variance with these factors (E
p
=
S
p
J
).
In addition to the dimensionality reduction which improves the efficiency of the
supervised learning algorithm, this process allows to detect and extract the true patterns
in the data, the last factors express the noisy information in the dataset. From this point
of view, the SVD is an effective data cleaning process, by selecting the p best factors,
we reject negligible information contained in the data. Thus, it is possible to reconstruct
an approximate version of original data from the selected factors and projection vectors.
About the implementation, the challenge was considerable. It was not possible to
use diagonalization techniques from the 8000 × 8000 correlation matrix in order to
extract eigenvalue and eigenvectors. It was thus necessary to consider the direct extrac-
tion of the singular values from the standardized matrix ℵ with a powerful algorithm,
the computing time and the memory requirement are major constraints. We used the NI-
PALS implementation [5] which interprets the singular value extraction as successive
orthogonal regressions: the first one produces the first factor F
1
, using the residuals of
this regression, we perform a new regression in order to produce the second factor F
2
,
etc. This approach allows to reduce computations considerably since we can stop calcu-
lations as soon as the first p factors were generated. In our experiments, from a n = 100
examples and J = 7000 descriptors, the first 5 factors are generated in 10 seconds on
a standard personal computer running under Windows (Pentium III – 1 Ghz – 512 MB
RAM). We use the TANAGRA [6], an open source data mining software, source code
is available on the website of the authors (http:\\eric.univ-lyon2.fr\˜ricco\tanagra).
2.2 SVD and Irrelevant Descriptors
If the SVD is a very interesting process for dimensionality reduction by controlling the
loss of information, it has a major drawback in a protein classification framework: the
SVD is an unsupervised process. In fact, to build the factors, it used all the descriptors,
including the irrelevant one for a supervised learning task.
40
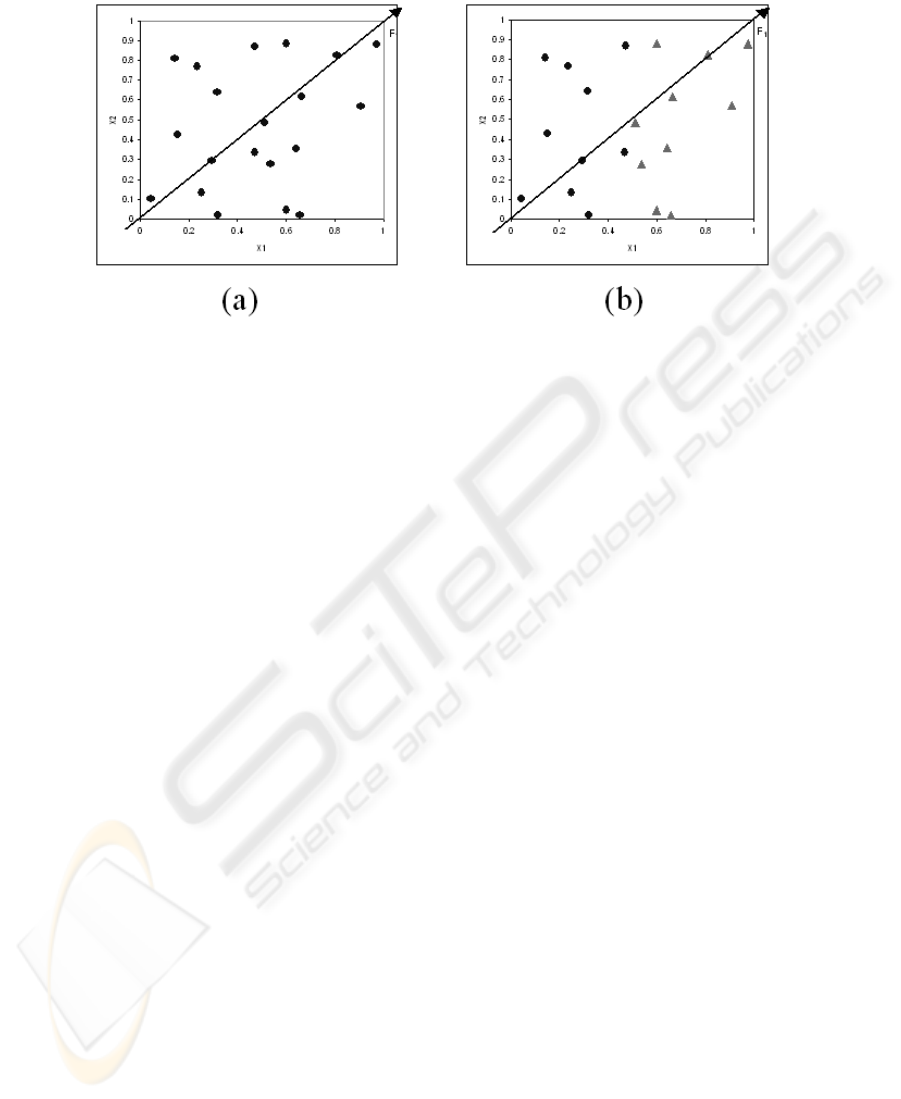
Fig.1. SVD on unsupervised (a) and supervised (b) tasks.
To illustrate this drawback, we show the same situation on an artificial two-dimen–
sional dataset (Figure 1). On the unlabeled dataset (Figure 1.a), the first extracted factor
F
1
seems appropriate, but on the labeled dataset (Figure 1.b), we see that the descriptor
X
2
is irrelevant for the learning task, however the SVD extracts the same factor F
1
.
In this paper, we propose to perform first a descriptor selection before building
the factors with SVD. We call this combination FS-SVD (Feature Selection - Singular
Value Decomposition). The goal of the selection is not to produce the most powerful
subspace for the prediction like in classical feature selection process [7] but rather to
eliminate the irrelevant descriptors before the SVD process. In this point of view, we
use a very simple filter algorithm: we rank the descriptors according to the correlation
coefficient criterion and keep the 50 best for SVD (the correlation coefficient computed
on 0/1 attribute is similar to χ
2
criterion on Boolean true/false attribute) [8]. Of course,
some selected descriptors are redundant but it is does not matter because the features
obtained with the SVD are orthogonal.
We propose the following framework for protein classification:
– Extract descriptors from native format of proteins sequences;
– Select the 50 most correlated descriptors with the class-attribute (protein family);
– Build and select the 5 first (best) features with the SVD process;
– Use these features in a supervised learning algorithm, we use a nearest neighbor
classifier (3-NN) because it is very sensitive to irrelevant descriptors and allows
us to evaluate our data preprocessing framework, especially the preservation of
the proximity between the examples [2]. We plan to test other supervised learning
algorithms in the near future.
The chosen parameters in our study (50 descriptors and 5 features) are defined in
an approximate way and are appropriate for all cases that we treated. Actually, in the
majority of cases, the first 2 factors are sufficient, but we preferred to make a simpli-
fied choice and avoid fine tuning parameters which is always problem dependent and a
41

Fig.2. Native description of proteins.
source of overfitting, especially when we use a cross validation error rate estimate. This
is particularly true in our case where we have a small number of examples compared to
a large number of descriptors.
3 Experiments on a Protein Classification Problem
3.1 The Protein Classification Problem
In this paper, we use the text mining framework for a protein classification problem
from their primary structures. The analogy with text classification is relevant in our case,
indeed the original description of the dataset is very similar. A protein is described by
a series of characters which represents amino acids. There are 20 possible amino acids.
We show an example of a file describing a few proteins (Figure 2).
However, unlike the text classification, there is no ”natural” separation in the char-
acter sequences, it is not possible to extract ”words” for which we can easily attach
semantics properties. Therefore, we have used the n-grams, a sequence of n consecu-
tive characters, in order to produce descriptors.
Previous works showed that the choice of n = 3 (3-grams) and boolean descriptors
give a good compromise to produce accurate classifier [1]. We obtain a Boolean at-
tribute - value dataset with several thousands of descriptors (Figure 3). The theoretical
maximum number of 3-grams for a protein classification problem is 20
3
= 8000. Of
course, all 3-grams are not present in a dataset but experiments showed that we were
close to this value. Numerous of 3-grams are irrelevant, others also are redundant. The
main challenge of the feature reduction is to build appropriate features for a supervised
learning task. There are several reasons for this dimensionality reduction: (1) machine
learning algorithms work badly when the dataset is too sparse; selecting a subset of
relevant features often improves the classifier performance; (2) the complexity of the
learning algorithms always depends on the number of input features; the elimination
of useless attributes allows to a considerable improvement in computing time; (3) a
reduced number of features provides a better understanding of the classifier.
42
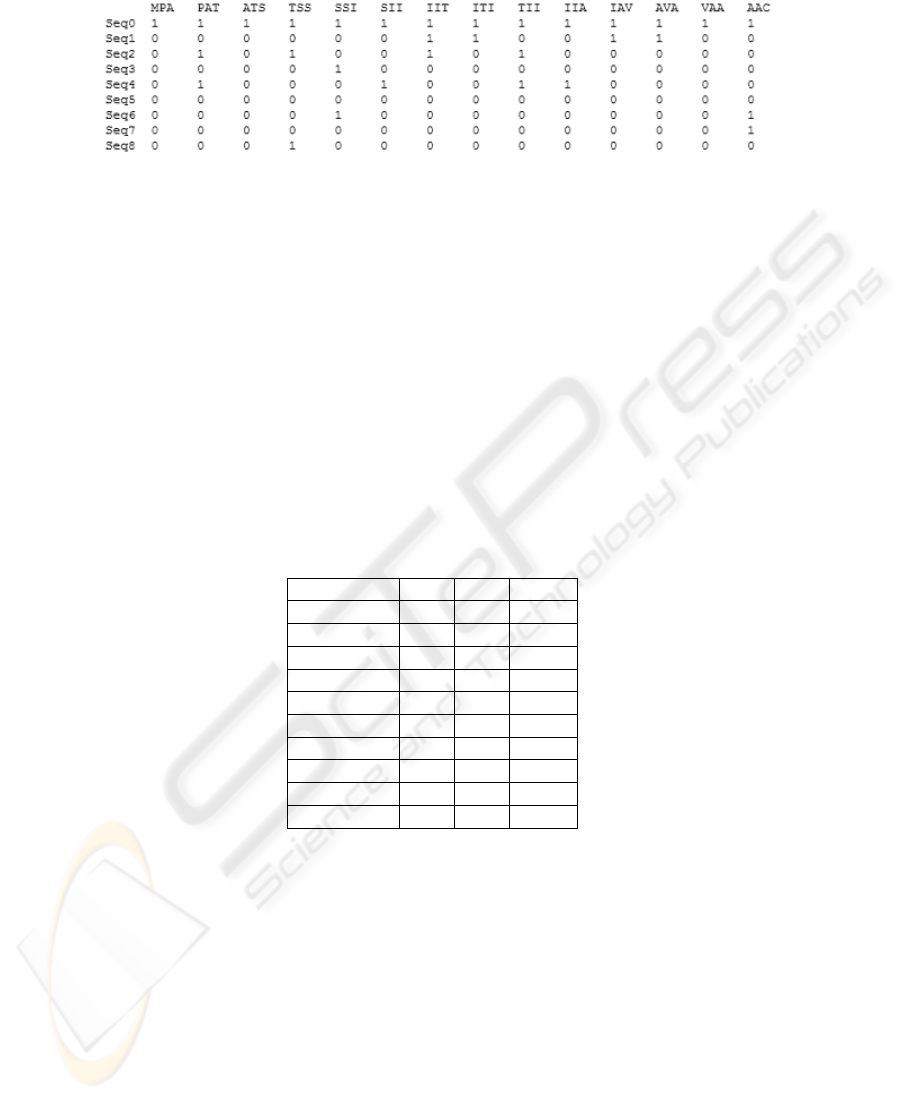
Fig.3. Boolean 3-grams attribute-value table from native description.
3.2 Experimental Results
Five protein families have been randomly extracted from the data bank SCOP [9], the
aim being to discriminate each pair of proteins. We use the bootstrap-plus error rate
estimate [10] instead of the standard cross-validation or leave-one out error rate estimate
because they can suffer of high-variability in certain cases.
In this paper, we compare 3 approaches: (ALL) we run the nearest neighbor al-
gorithm on all descriptors; (SVD) we run the learning algorithm on the 5 first factors
extracted from all descriptors; (FS-SVD) we perform a selection of 50 best descriptors
and run the learning algorithm on the 5 first factors extracted using a SVD from these
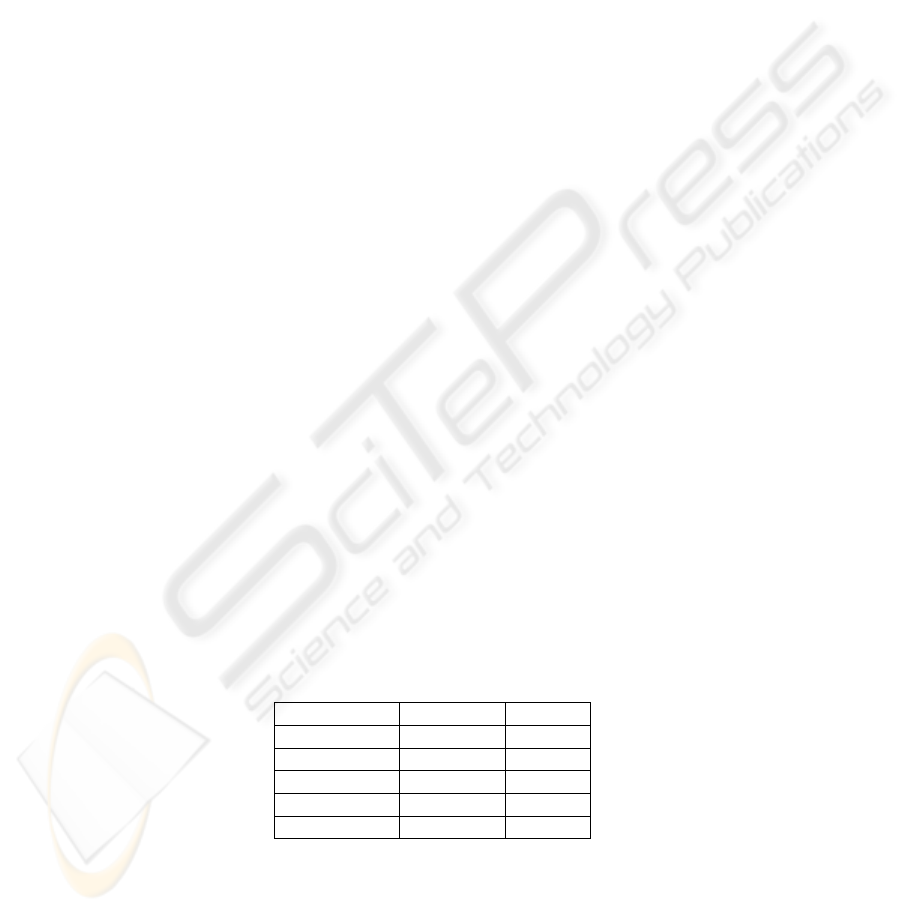
descriptors. The results are available in Table 1.
Table 1. Estimated error rate on each protein discrimination problem.
Proteins pairs ALL SVD FS-SVD
F
12
0.1778 0.0445 0.0024
F
13
0.1728 0.0015 0.0045
F
14
0.1293 0.0189 0.0035
F
15
0.1664 0.0083 0.0019
F
23
0.2593 0.1653 0.0308
F
24
0.1113 0.0731 0.0276
F
25
0.1496 0.1301 0.0213
F
34
0.2073 0.0805 0.0310
F
35
0.2328 0.1111 0.0568
F
45
0.1441 0.1685 0.0387
The results suggest some interesting comments:
– Running the learning algorithm on all descriptors is an inefficient approach, the
high dimensionality deteriorates the results, because there are many irrelevant de-
scriptors, but also because the nearest neighbor works poorly when we have a sparse
dataset.
– The SVD approach improves the performance of the classifier, the dimensionality
reduction allows the learning algorithm to work well. Let us note that in the ma-
jority of the problems studied, the first 5 factors restore 15% of the information
43
contained in the data. We will further study below the influence of the number of
selected factors on the performance of the classifier.
– Descriptor selection before building the factors is an efficient way to improve the
classifier performance. In all cases, FS-SVD outperforms SVD, removing irrelevant
descriptors helps the singular value decomposition technique to build more relevant
features (factors) for the learning algorithm.
Even if that were not our first goal in this work, it nevertheless were interesting
to compare the computing times between the two approaches (SVD and FS-SVD): we
noted that, on average, the descriptor selection allows to reduce 15 times the execution
time of the protein classification problems.
4 Discussion: Further Experiments
4.1 The Influence of the Number of Factors
On the FS-SVD process, a detailed study of the results showed that very often the first
2 factors are sufficient to produce powerful classifier. We can use a feature selection
process to individually evaluate each factors. Because they are orthogonal, an evaluation
of their relevance can be independently used but we think that it is not decisive in our
case, this is why we make the default choice of 5 factors.
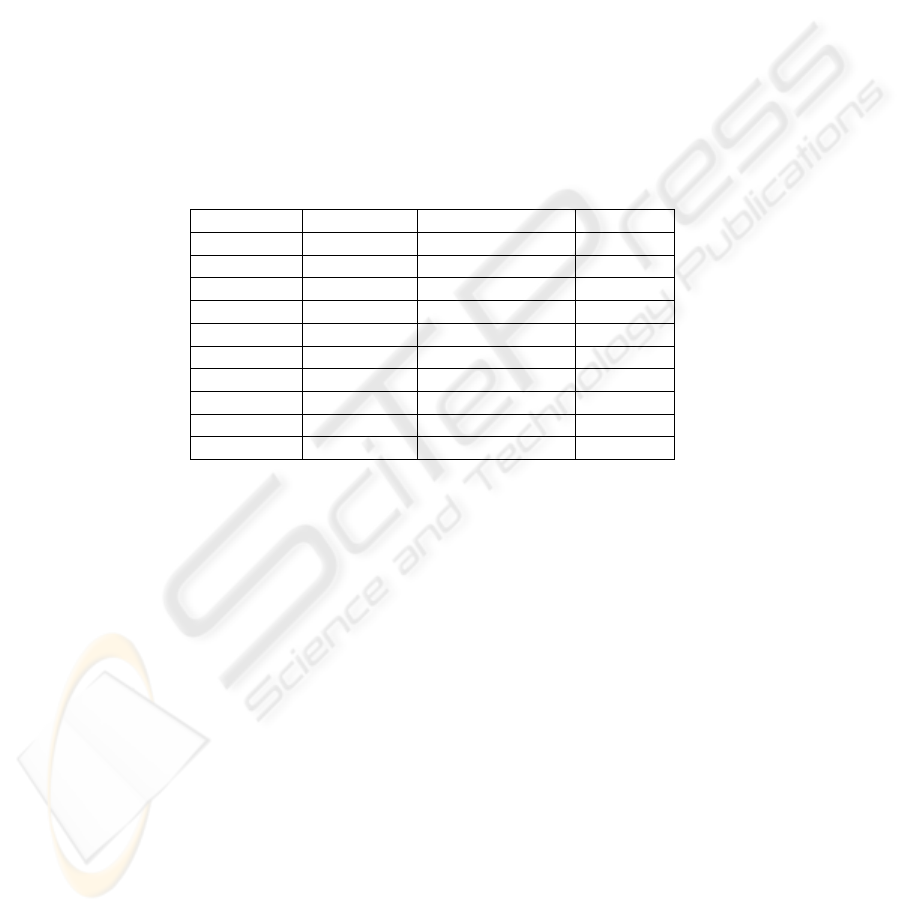
More interesting was the choice of the number of factors for the SVD process. The
choice of 5 factors allows us to compare the two approaches but we have seen that in
this case, the loss of information is nevertheless significant. For the F
34
problem (Table
1) , we set several values of extracted factors with the SVD approach, we measure the
explained variance and the error rate of the learning classifier (Table 2). We see that the
trade-off between the quality of the representation (explained variance on the selected
factors) and efficiency of the learning algorithm (which suffers of the increase of the
representation space) is not easy to find. To introduce a fine adjustment of the number
of selected factors in order to optimize the error rate is at the opposite of our approach,
moreover that would increase the risks of overfitting.
Table 2. Explained variance and error rate of the classifier for the F
34
problem.
Selected factors (%) variance Error rate
5 15 0.0805
10 24 0.0801
20 38 0.1100
50 67 0.2063
100 95 0.2129
44
4.2 Feature Construction vs. Feature Selection
Select relevant and non redundant features leads to improved classification accuracy.
In this paper, the descriptor ranking allows to eliminate irrelevant descriptors, the SVD
process allows to build orthogonal features from the relevant descriptors.
In order to improve the classifier performance, another solution is to perform a more
aggressive descriptor selection which combines the detection of the relevance and the
elimination of redundancy. The correlation based approach seems a promising way in
this domain, especially in the microarray data analysis. The FCBF method [11] can
make the best trade-off between relevance and redundancy. It is important in this paper
to verify, in the first time, if this approach leads to better classifier, and in the second
time, to consider the respective advantage of the two approaches, feature construction
from SVD and redundancy based descriptor selection.
Table 3. Number of the selected descriptors and error rate with FCBF. Comparison with FS-SVD.
Proteins pairs Err. (FS-SVD) Descriptors (FCBF) Err. (FCBF)
F
12
0.0024 28 0.0048
F
13
0.0045 30 0.0022
F
14
0.0035 30 0.0028
F
15
0.0019 23 0.0020
F
23
0.0308 5 0.0476
F
24
0.0276 13 0.0272
F
25
0.0213 6 0.0702
F
34
0.0310 9 0.0376
F
35
0.0568 6 0.0649
F
45
0.0387 12 0.0262
Roughly speaking, FCFB ranks the features using a correlation measurement. It
selects a feature (1) if its correlation with class attribute is upper than δ (a parameter
of the algorithm); (2) if it is predominant i.e. its correlation with the class attribute is
upper than its cross-correlation to the all other features. In our experiment, it is clear that
FCBF is heavily parameter dependent. The δ parameter which allows to control the size
of selected descriptors subset is very hard to adjust. We use the standard δ = 0.3 which
seems a good compromise for our files, experiments results are reported in Table 3. It
seems that FCBF gives similar results to our approach, in 4 dataset FCBF outperforms
FS-SVD, the dimensionality reduction is effective. It is even possible to obtain better
results by adjusting the parameter; in this case, the number of selected descriptors can
be modified without significant improvement of the classifier performance.
But a detailed study of the results calls into question theses results. Indeed, in our
dataset the descriptors are automatically generated from an unstructured data format,
the choice of 3-grams is a compromise in order to obtain a efficient classifier, the elim-
ination of the redundant descriptors is a purely mechanical process, it masks the con-
comitant action of two or several descriptors. For instance, for the discrimination of
45

”Tool Like Receptor” protein family, experts know that the 4-gram ”LDLS” is a signifi-
cant descriptor. Because we use 3-grams, we obtain ”LDL” and ”DLS”, the redundancy
based methods eliminates one of them, thus preventing any thorough interpretation of
the results.
The SVD offers several kind of visualization of the results. Its advantages in protein
discrimination are interesting: we can at the same time study our data as well from the
point of view of the coordinate of individuals in the new representation space, as the
point of view of the evaluation of the influences of the descriptors in the construction
of the ”latent variables”. This ”pattern detection” property of SVD can be very useful
in the search of more powerful and interpretable descriptors than the simple 3-grams.
We can for instance build a 4-grams from 2 compatibles 3-grams which are highly
correlated with the first factor. We manually did it for the moment, but it appears that it
is a promising approach if we find a strategy to automatize this process.
5 Conclusion
In this paper we show that elimination of irrelevant descriptors allows the singular value
decomposition to produce more efficient factors for the protein classification context
where we have a high dimensional boolean dataset. The classifier accuracy is improved
significantly. In the same time, the computing time is dramatically reduced. Our ap-
proach is rather robust because we can avoid any fine tuning of the parameters, the risks
of overfitting are reduced.
These results open new perspectives. Indeed, the singular value decomposition of-
fers powerful tools for interpretation of results which make it possible for the expert to
improve his knowledge of the domain and to propose some explorations which can, in
particular, lead to the creation of more powerful and understandable descriptors.
In this paper, we see that combining feature selection and SVD allows to increase
the performances of K-NN which is very sensitive to high dimensionality. In a future
work, it will be interesting to study the behavior of this data processing on more robust
learning algorithms such as linear support vector machine and try to characterize the
context where this approach is the most powerful.
References
1. Mhamdi, F., Elloumi, M., Rakotomalala, R.: Text-mining, feature selection and data-mining
for proteins classification. In: Proceedings of International Conference on Information and
Communication Technologies: From Theory to Applications, IEEE Press (2004) 457–458
2. Hastie, T., Tibshirani, R., Friedman, J.: The Elements of Statistical Learning: Data Mining,
Inference, and Prediction. Springer (2001)
3. Wall, M., Rechtsteiner, A., Rocha, L.: Singular Value Decomposition and Principal Com-
ponent Analysis. In: A Practical Approach to Microarray Data Analysis. Kluwer (2003)
91–109
4. Husbands, P., Simon, H., Ding, C.: On the use of the singular value decomposition for
text retrieval. In: Proceedings of 1st SIAM Computational Information Retrieval Workshop.
(2000)
46

5. Wold, S., Esbensen, K., Geladi, P.: Pricnipal component analysis. Chemometrics and Intel-
ligent Laboratory Systems 2 (1987) 37–52
6. Rakotomalala, R.: Tanagra: une plate-forme d’exp
´
erimentation pour la fouille de donn
´
ees.
Revue MODULAD (2005) 70–85
7. Guyon, I., Elisseeff, A.: An introduction to variable and feature selection. Journal of Machine
Learning Research 3 (2003) 1157–1182
8. Duch, W., Wieczorek, T., Biesiada, J., Blachnik, M.: Comparison of feature ranking methods
based on information entropy. In: Proceedings of International Joint Conference on Neural
Networks (IJCNN), IEEE Press (2004) 1415–1420
9. Murzin, A., Brenner, S., Hubbard, T., Chothia, C.: Scop: a structural classification of proteins
database for the investigation of sequences and structures. Journal of Molecular Biology
(1995) 536–540
10. Efron, B., Tibshirani, R.: Improvements on cross-validation: The 0.632+ bootstrap method.
JASA 92 (1997) 548–560
11. Yu, L., Liu, H.: Redundancy based feature selection for microarray data. In: KDD ’04:
Proceedings of the 2004 ACM SIGKDD international conference on Knowledge discovery
and data mining, New York, NY, USA, ACM Press (2004) 737–742
47
