
Colorectal Cancer Classification using Deep Convolutional Networks
An Experimental Study
Francesco Ponzio, Enrico Macii, Elisa Ficarra and Santa Di Cataldo
Department of Control and Computer Engineering, Politecnico di Torino, Corso Duca degli Abruzzi 24, 10129 Torino, Italy
Keywords:
Colorectal Cancer, Histological Image Analysis, Convolutional Neural Networks, Deep Learning, Transfer
Learning, Pattern Recognition.
Abstract:
The analysis of histological samples is of paramount importance for the early diagnosis of colorectal cancer
(CRC). The traditional visual assessment is time-consuming and highly unreliable because of the subjectivity
of the evaluation. On the other hand, automated analysis is extremely challenging due to the variability of
the architectural and colouring characteristics of the histological images. In this work, we propose a deep
learning technique based on Convolutional Neural Networks (CNNs) to differentiate adenocarcinomas from
healthy tissues and benign lesions. Fully training the CNN on a large set of annotated CRC samples provides
good classification accuracy (around 90% in our tests), but on the other hand has the drawback of a very
computationally intensive training procedure. Hence, in our work we also investigate the use of transfer
learning approaches, based on CNN models pre-trained on a completely different dataset (i.e. the ImageNet).
In our results, transfer learning considerably outperforms the CNN fully trained on CRC samples, obtaining
an accuracy of about 96% on the same test dataset.
1 INTRODUCTION
Colorectal carcinoma (CRC) is one of the most dif-
fused cancers worldwide and one of the leading
causes of cancer-related death. According to recent
epidemiological data, this type of cancer has signifi-
cant burden in most of the European countries, and it
is still associated with very high mortality rates (Mar-
ley and Nan, 2016). Hence, the early diagnosis and
differentiation of the tumour is crucial for the survival
and well-being of a large number of patients.
Traditionally, pathologists perform CRC diagno-
sis by visually examining under the microscope the
resected tissue samples, fixed and stained by means
of Hematoxylin and Eosin (H&E). The presence and
level of malignancy is assessed by observing the or-
ganisational changes in the tissues, which are high-
lighted by the two stains. As shown in Figure 1, nor-
mal colon tissues have a well-defined organisation,
with the epithelial cells forming glandular structures
and the non-epithelial cells (i.e. stroma) lying in be-
tween these glands. The main benign precursor of
CRC, adenoma, is characterised by enlarged, hyper-
chromatic and elongated nuclei arranged in a typically
stratified configuration. Compared to normal tissues,
the adenoma is characterised by either tubular or vil-
lous (finger-like) tissue architecture. Adenocarcino-
mas, on the other hand, produce abnormal glands that
infiltrate into the surrounding tissues.
As it is widely pointed out by literature, manual
examination has two major drawbacks. First, it is
time-consuming, especially for large image datasets.
Figure 1: Histological H&E images of colorectal tissues
(cropped patches). i) Healthy tissue; ii) Adenocarcinoma;
iii) Tubulovillous adenoma.
Second, it is highly subjective and affected by
58
Ponzio, F., Macii, E., Ficarra, E. and Cataldo, S.
Colorectal Cancer Classification using Deep Convolutional Networks - An Experimental Study.
DOI: 10.5220/0006643100580066
In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2018) - Volume 2: BIOIMAGING, pages 58-66
ISBN: 978-989-758-278-3
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

variability, both inter and intra observer (A. Young
and Kerr, 2011). Hence, there are growing efforts
towards the development of computer-aided diagnos-
tic techniques, with two major directions: (i) auto-
mated segmentation, aimed at partitioning the hetero-
geneous colorectal samples into homogeneous (i.e.
containing only one type of tissue) regions of inter-
est. (ii) automated classification, aimed at categoris-
ing the homogeneous tissue regions into a number of
classes, either normal or malignant, based upon quan-
titive features extracted from the image. In both the
tasks, the main challenge to be tackled is the extreme
intra-class and inter-dataset variability, that is an in-
herent characteristic of histological imaging. In this
work, we focus on the automated classification task,
and specifically into three histological categories that
are most relevant for CRC diagnosis: (i) healthy tis-
sue, (ii) adenocarcinoma, (iii) tubulovillous adenoma.
In the last few years, the literature on automated
classification of histological images has been exten-
sive, with applications covering different anatomical
parts other than colon, such as brain, breast, prostate
and lungs. Most of the proposed approaches rely on
automated texture analysis, where a limited set of
local descriptors are computed from patches of the
original input images and then fed into a classifier.
Among the most frequently used, statistical features
based on grey level co-occurrence matrix (GLCM),
local binary patterns (LBP), Gabor and wavelet trans-
forms, etc. The texture descriptors, eventually en-
coded into a compact dictionary of visual words, are
used as input of machine learning techniques such
as Support Vector Machines (SVM), Random Forests
or Logistic Regression classifiers (Di Cataldo and Fi-
carra, 2017). In spite of the good level of accuracy
obtained by some of these works, the dependence on
a fixed set of handcrafted features is a major limita-
tion to the robustness of the classical texture analysis
approaches. First, because it requires a deep knowl-
edge of the image characteristics that are best suited
for classification, which is not obvious. Second, be-
cause it puts severe constraints to the generalisation
and transfer capabilities of the proposed classifiers,
especially in presence of inter-dataset variability.
As an answer to such limitations, in the recent
years the use of deep learning (DL) architectures,
and more specifically Convolutional Neural Networks
(CNNs), has become a major trend (Janowczyk and
Madabhushi, 2016; Korbar et al., 2017). In CNNs
a number of convolutional and pooling layers learns
by backpropagation a set of features that are best for
classification, thus avoiding the extraction of hand-
crafted texture descriptors. Nonetheless, the necessity
of training the networks with a huge number of in-
dependent histological samples is still an open issue,
which limits the usability of the approach in the ev-
eryday clinical setting. Transfer learning (i.e applying
CNNs pre-trained on a different type of images, for
which large datasets are available) seems a promising
solution to this problem (Weiss et al., 2016) but not
fully investigated for CRC classification.
In this work. we evaluate a CNN-based ap-
proach to automatically differentiate healthy tissues
and tubulovillous adenomas from cancerous samples,
which is a challenging task in histological image anal-
ysis. For this purpose, we fully train a CNN on a large
set of colorectal samples, and assess its accuracy on
an independent test set. This technique is experimen-
tally compared with two different transfer learning
approaches, both leveraging upon a CNN pre-trained
on a completely different image dataset. The first ap-
proach uses the pre-trained CNN to extract a set of
discriminative features that will be fed into a sepa-
rate Support Vector Machines classifier. The second
approach fine-tunes on CRC histological images only
the last stages of the pre-trained CNN. By doing so,
we investigate and discuss the transfer learning capa-
bilities of CNNs in the domain of colorectal tissues
classification.
2 MATERIALS AND METHODS
2.1 Colorectal Cancer Image Dataset
The dataset used in this study was extracted from
a public repository of H&E stained whole-slide im-
ages (WSIs) of colorectal tissues, available on line
at http://www.virtualpathology.leeds.ac.uk/. All the
slides are freely available for research purposes, to-
gether with their anonymised clinical information.
In order to obtain a statistically significant dataset
in terms of inter-subjects and inter-class variability,
27 WSIs were selected among univocal subjects (i.e.
one WSI per patient). Note that different types of tis-
sues (e.g. healthy and cancerous portions) coexist in a
single WSI. With the supervision of a skilled pathol-
ogist, we identified large regions of interest (ROIs)
on the WSIs as in the example of Figure 3, so that
each ROI is univocally associated to one out of the
three tissue subtypes: (i) adenocarcinoma (AC); (ii)
tubuvillous adenoma (TV) and (iii) healthy tissue (H).
Then, the ROIs were cropped into a total number of
13500 1089x1089 patches (500 per patient), at a mag-
nification level of 40x.
For training and testing purposes, the original
image cohort was randomly split into two disjoint
subsets, comprising 18 subjects for training (9000
Colorectal Cancer Classification using Deep Convolutional Networks - An Experimental Study
59

Figure 2: CNN architecture.
patches in total) and 9 for testing (4500 patches). See
Table 1 for a complete characterisation of the two sets.
The random sampling was stratified, so that both the
training and the testing dataset are balanced among
the three classes of interest (H, AC and TV, respec-
tively).
Table 1: CRC image dataset.
Train Test Tot
# of patients 18 9 27
# of ROIs 85 24 109
# of patches 9000 4500 13500
Before being fed into the CNN, each patch was
down sampled by a factor five, which was empirically
set as a trade-off between computational burden of the
processing and architectural detail of the images. In
order to compensate for the color inconsistencies, the
patches were normalised by mean and standard devi-
ation, computed over the whole training dataset.
2.2 Convolutional Neural Network:
Architecture and Training
Paradigm
A Convolutional Neural Network (CNN) is made up
of multiple locally connected trainable stages, piled
one after the other, with two or more fully-connected
layers as the last step. The first part of the network
Figure 3: Identification of ROIs from a WSI: example.
is devoted to learning the image representation, with
successive layers learning features at a progressively
increasing level of abstraction, while the last fully-
connected part is devoted to classification and acts
like a traditional multilayer perceptron.
From a computational point of view, a CNN archi-
tecture is characterised by two main types of building
blocks:
(i) Convolutional (CONV) blocks, that perform
a 2D convolution operation (i.e. kernel filter-
ing) on the input image and apply a non-linear
transfer function, such as Rectified Linear Unit
(ReLU). Based on the trainable parameters of
the kernels, the stage detects different types of
local patterns on the input image.
(ii) Pooling (POOL) blocks, that perform a non-
linear down-sampling of the input (e.g. by ap-
plying a max function). This has the double ef-
fect of reducing the amount of parameters of the
network to control overfitting and of making the
image representation (i.e. the local pattern de-
scriptors learnt by the network) spatially invari-
ant.
The number of CONV and POOL blocks (i.e. the
depth) of the network is directly related to the level
of detail that can be achieved in the the hierarchical
representation of the image. Nonetheless, a higher
depth also translates into a higher number of parame-
ters, and hence on a higher computational cost.
The training paradigm chosen for the CNN is
a classic backpropagation scheme: an iterative pro-
cess that involves multiple passes of the whole input
dataset until the model converges. At each training
step, the whole dataset flows from the first to the last
layer in order to compute a classification error, quan-
tified by a loss function. Such error flows backward
through the net, and at each training step the model
parameters (i.e. the network weights) are tuned in the
direction that minimises the classification error on the
training data.
As a trade-off between representation capabili-
BIOIMAGING 2018 - 5th International Conference on Bioimaging
60

ties and computational costs, in our work we used
a VGG16 CNN model, which is represented in Fig-
ure 2 (Simonyan and Zisserman, 2014). This ar-
chitecture was successfully applied to a large num-
ber of computer vision tasks. In spite of the quite
large depth, the VGG16 adopts a very simple archi-
tecture, based on piling up only 3x3 convolution and
2x2 pooling blocks. More specifically, the model con-
sists of 13 CONV layers that can be conceptually
grouped into 5 macro-blocks ending with one POOL
layer each, and of a final 3-layered fully-connected
(FC) stage. Non-linearities are all based on ReLU,
except for the last fully-connected layer (FC3), that
has a softmax activation function. The convolution
stride and the padding are fixed to 1 pixel and the max
pooling stride to 2. Differently from the original ar-
chitecture of VGG16, in our work the size of FC3 is
3, matching the number of categories targeted by our
research problem.
The net was built within Keras framework (Chol-
let et al., 2015) and trained with a backpropagation
paradigm. More specifically, we applied a stochastic
gradient descent (SGD), implemented with a momen-
tum update approach (Qian, 1999) as iterative opti-
misation algorithm to minimise the categorical cross-
entropy function between the three classes of interest
(H, AC and TV). To monitor the training and optimise
the choice of hyper-parameters of the net, we used
10% of the training set as validation data. This sub-
set is completely independent from the images used
for testing purposes, and was solely used to compute
the validation accuracy metric upon which the train-
ing process is optimised. Based on validation, we se-
lected a learning rate (LR) of 0.0001, a momentum
(M) of 0.9 and a batch size (BS) of 32 images. The
learning strategy involved the so-called early stopping
(i.e., the training is stopped when validation accuracy
does not improve for 10 subsequent epochs), as well
as the progressive reduction of LR each time the val-
idation accuracy does not improve for 5 consecutive
epochs. Such technique was found to largely reduce
overfitting (Yao et al., 2007).
The CNN was trained for 30 epochs on our col-
orectal cancer training dataset, which lasted 8 hours
on Linux Infiniband-QDR MIMD Distributed Shared-
Memory Cluster provided with single GPU (NVIDIA
Tesla K40 - 12 GB - 2880 CUDA cores). Figure 4
shows the loss (a) and accuracy (b) curves on both the
training and validation datasets.
From the graphs of Figure 4 we can derive the fol-
lowing observations:
(i) The model seems to converge quite quickly. In-
deed, while training accuracy is still increasing,
the value of validation accuracy saturates within
15 epochs.
(ii) The decay speed of the validation loss curve in-
dicates that the learning rate is appropriate.
(iii) The similarity of validation and training accura-
cies reasonably rules out overfitting.
2.3 Transfer Learning From
Pre-trained CNN
A CNN is a cascade of trainable filter banks, where
the first blocks of filters are devoted to the detection
of low-level features (i.e. edges or simple shapes),
and the following ones are activated by high-level se-
mantic aggregations of the previous patterns. While
the top-most blocks are generally tailored to a specific
classification task, the lower-level features are ideally
generalisable to a large number of applications. This
concept, that is at the basis of all the transfer learning
techniques using CNNs as feature generators, lever-
age on the assumption that the network had first been
trained on a very large set of examples, with signifi-
cant variability of image characteristics.
In our work, we performed experiments using a
pre-trained CNN model with the same architecture
and building blocks of the one used for full train-
ing on colorectal cancer images (VGG16, shown in
Figure 2). The model was pre-trained on the Ima-
geNet dataset, from the Large Scale Visual Recog-
nition Challenge 2012 (ILSVRC-2012). The Ima-
Figure 4: Training vs validation loss per epoch (a) and train-
ing vs validation accuracy per epoch (b).
Colorectal Cancer Classification using Deep Convolutional Networks - An Experimental Study
61
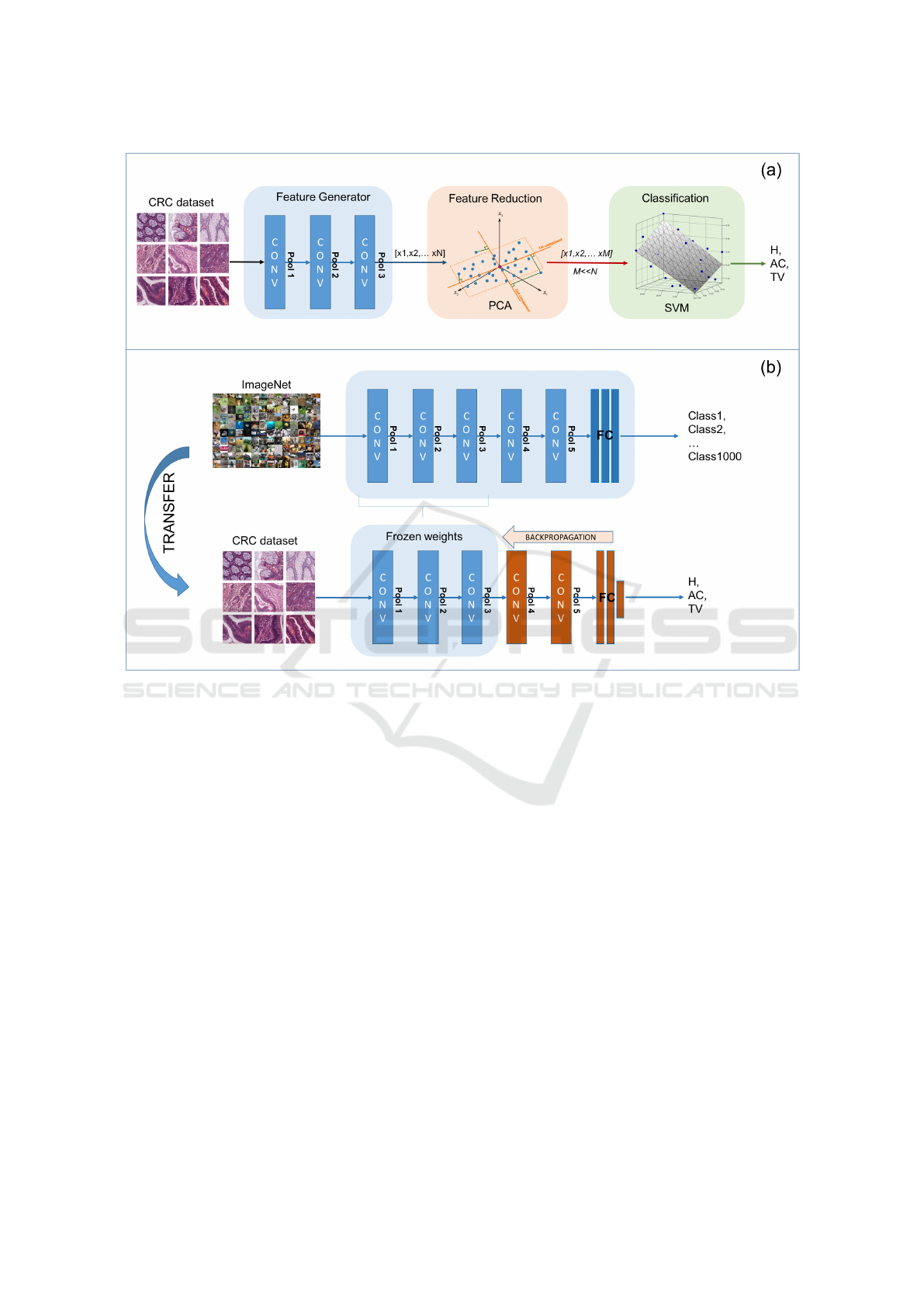
Figure 5: Transfer learning approaches. (a) Pre-trained CNN as a fixed feature generator. (b) Fine tuning of pre-trained CNN.
geNet dataset contains 1.2 million photographs de-
picting 1000 different categories of natural objects.
Hence, the content and characteristics of the training
images are completely different from our specific tar-
get.
To apply the pre-trained CNN to our histology
classification task, we implemented and compared
two different transfer learning approaches, whose
main steps are represented in Figure 5 (a) and (b), re-
spectively:
(i) CNN as a fixed feature generator. The his-
tological images are given as input to the pre-
trained CNN for inference. The features ex-
tracted by the convolutional blocks are then fed
into a separate machine learning framework,
consisting of a feature reduction stage and a su-
pervised classifier.
(ii) Fine-tuning the CNN. The CNN model is re-
trained on our training set of histological im-
ages, keeping all the parameters of the low-level
blocks fixed to their initial value. Hence, only
the weights of the top-most layers are fine-tuned
for colorectal cancer classification.
As a preliminary step to both the two approaches,
we analysed the discriminative capabilities of the fea-
tures generated by all the major blocks of the pre-
trained CNN. More specifically, we randomly se-
lected a small subset of the training images (i.e. 1500
patches, 500 per class) and we fed these images into
the pre-trained CNN. The output of each successive
macro-block of the CNN was then analysed, to assess
the degree of separation of samples belonging to the
three different classes. As a trade-off between thor-
oughness and computational burden of the investiga-
tion, we analysed the intermediate output of the CNN
only at the end of the pooling layers (i.e. POOL1 to
5, in Figure 2). Indeed, as the pooling layers perform
a feature reduction on the output of the convolutional
filters, they are expected to produce a non-redundant
set of image features compared to CONV layers.
The degree of class separation was assessed by
means of t-Distributed Stochastic Neighbour Embed-
ding (t-SNE) (Maaten and Hinton, 2008), a non-linear
dimensionality reduction algorithm that is used for
BIOIMAGING 2018 - 5th International Conference on Bioimaging
62
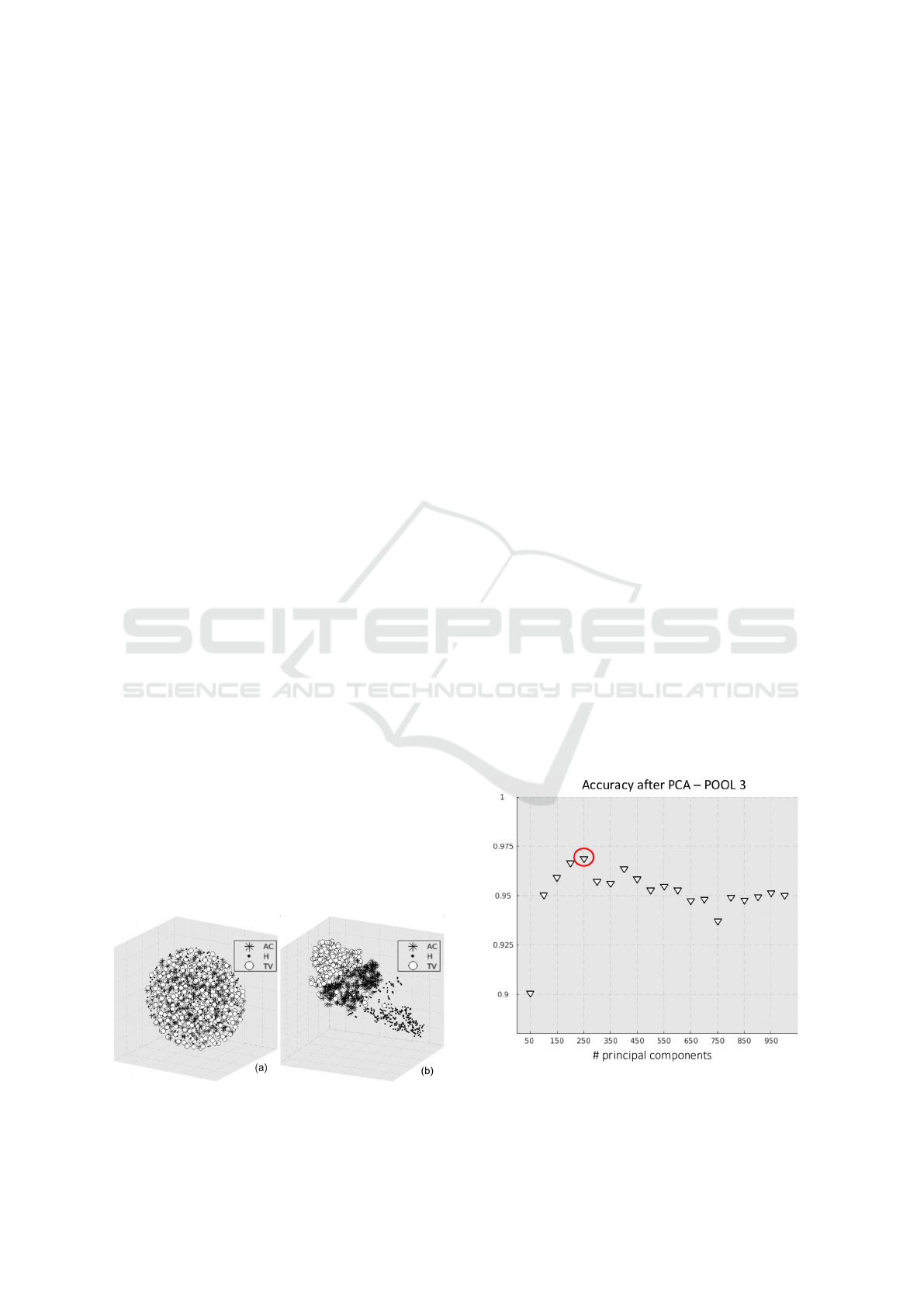
the visualisation of high-dimensional datasets in a re-
duced 3-dimensional space. More specifically, t-SNE
models each high-dimensional object (in our case, the
feature vector obtained at the output of a POOL layer)
by means of or three-dimensional point in a cartesian
space, so that similar feature vectors are represented
by nearby points and dissimilar vectors by distant
points. This allows to qualitatively assess the class
separability in the original feature space, and hence to
establish the POOL block that ensures the best class
separability (see examples in Figure 6).
The outcome of t-SNE was confirmed by further
quantitative experiments, based on assessing the clas-
sification performance of a separate classifier trained
on different POOL blocks. In all such experiments,
POOL3 outperformed all the other blocks.
2.3.1 Pre-trained CNN as A Fixed Feature
Generator
As first transfer learning methodology, the output of
the most discriminative POOL layer of the pre-trained
CNN (in our case POOL3) was used to generate
a feature vector for colorectal cancer classification.
The feature vector was fed into the machine learning
framework represented by Figure 5-(a), consisting in
a feature reduction and a classification step.
(i) Feature reduction. Principal Component Anal-
ysis (PCA) was applied to reduce the dimen-
sionality of the input data and prevent overfit-
ting. PCA performs an orthogonal transforma-
tion of the original features into the so-called
principal components, a new group of values
which are linear combinations of the original
characteristics. As PCA works towards the min-
imisation of the correlation between the fea-
tures, the new data representation is expected to
best summarise those features which are most
representative for the classes of interest. In our
work, the optimal number of principal compo-
nents was empirically determined by means of
a sequential forward procedure. The mean clas-
Figure 6: t-SNE visualisation of the output of POOL2 (a)
and POOL3 (b).
sification accuracy obtained on the training set
was computed at increasing number of principal
components, with a step of 50. To limit the com-
putational cost of the procedure, we selected the
minimum number of principal components af-
ter which the classification accuracy had started
decreasing, that was equal to 250 (see Figure 7).
(ii) Classification. The final classification into
three categories (H, AC, TV) was performed by
a Support Vector Machine (SVM) with a Gaus-
sian radial basis function kernel. The hyper-
parameters of the kernel were set by means of a
Bayesian Optimisation (BO) algorithm (Hastie
et al., 2009), implementing a 10-fold cross-
validation procedure on the training images. BO
was found to provide much better and faster re-
sults compared to classic methods based on grid
search or heuristic techniques.
2.3.2 Fine-tuning of Pre-trained CNN
As a second transfer learning methodology, we tried
to adapt the pre-trained VGG16 net to our specific
classification task. For this purpose, we first ini-
tialised all the weights of the network to the ones
determined on the ImageNet dataset, as represented
in Figure 5-(b). Then, we continued the backprop-
agation procedure on our CRC dataset, keeping the
weights of the first blocks of the net frozen. More
specifically, we froze all the weights up-to the most
discriminative pooling layer (POOL3), as determined
by t-SNE in Section 2.3. The rationale of such strat-
egy is trying to maintain the low-level features de-
scribing the most generic and generalisable details
Figure 7: Sequential forward procedure to select the optimal
number of principal components for PCA.
Colorectal Cancer Classification using Deep Convolutional Networks - An Experimental Study
63

(e.g. edges and simple shapes) as they were learnt
from the ImageNet. Hence, all the computational
power can be devoted to the training of the top-most
layers, which are expected to learn high-level task-
specific features for colorectal image classification.
The training strategy was exactly the same that was
described in Section 2.2.
3 CLASSIFICATION ACCURACY
3.1 Performance Metrics
The classification performance was assessed using the
dataset described in Section 2.1. As already pointed
out, the test dataset is completely independent from
the one used for training the network and optimising
the classification parameters. The accuracy of the sys-
tem was assessed at two different levels of abstraction
(per patch and per patient, respectively). For this pur-
pose, we introduce two different performance metrics.
(i) Patch score: (S
P
), defined as the fraction of
patches of the test set that were correctly clas-
sified:
S
P
=
N
C
N
,
where N
C
is the number of correctly classified
patches and N the total number of patches in the
test set.
(ii) Patient score: (S
Pt
), defined as the fraction of
patches of a single patient that were correctly
classified (i.e. per-patient patch score), aver-
aged over all the patients in the test set:
S
Pt
=
∑
i
S
P
(i)
N
P
,
where S
P
(i) is the patch score of the i − th pa-
tient and N
P
the total number of patients in the
test set.
3.2 Results and Discussion
In Table 2 we report both the patch and patient scores
obtained for the three classification frameworks de-
scribed in Section 2. More specifically:
(i) full-train-CNN refers to the CNN fully trained
on CRC samples.
(ii) CNN+SVM: refers to the SVM, with pre-
trained CNN used as fixed feature generator.
(iii) fine-tune-CNN: refers to the pre-trained CNN
with fine-tuning of the final stages.
For the patient score, S
Pt
value is reported as
mean ± standard deviation. From the results of
Table 2 we can observe that all the proposed clas-
sification frameworks obtained accuracy (both patch
and patient-wise) above 90%. Hence, the promising
results obtained by CNNs in other contexts are con-
firmed even for the application targeted by our work.
On top of that, the accuracy computed over all the
patches of the test set (S
P
) is very similar to the one
computed patient per patient (S
Pt
), with a very small
standard deviation of the latter value. This suggests
that the classification frameworks are all quite robust
and cope well with inter-patient variability, that is a
typical challenge of histopathological image analysis.
The same conclusions hold if we analyse the per-
class results, that are reported in the form of 3X3 con-
fusion matrices in Figure 8. In addition, from the
confusion matrices we can observe that the perfor-
mance of the classification frameworks is fairly ho-
mogeneous for the three classes H, AC and TV.
Quite interestingly, both the methodologies based
on transfer learning overcome the accuracy obtained
by the CNN fully trained on colorectal samples by al-
most 7%. In particular, the approach that provided
the best accuracy values (both patch and patient-
wise) was the pre-trained CNN with fine-tuning of the
blocks following POOL 3. This suggests the follow-
ing:
(i) Even though the full training seemed to con-
verge well and without overfitting on the train-
ing images (see Figure 4), the CNN would prob-
ably necessitate a much larger cohort of exam-
ples to learn features that are sufficiently general
to cope with the high variability of histopatho-
logical images. On the other hand, much larger
training datasets would make the learning pro-
cess prohibitive, especially in a clinical context.
(ii) In spite of the fact that the pre-training was
performed on a completely different dataset
(i.e. the ImageNet, which contains photographs
of every-day objects and natural scenes, and
not histological samples), the low-level features
learnt by the first stages of a CNN can be suc-
cessfully generalised to the context of CRC im-
age classification. Hence, CNNs are as a mat-
ter of fact capable of extracting usable seman-
tic knowledge from totally different domains.
This is very encouraging, as it partially avoids
Table 2: Patch and patient scores on the test set.
S
P
S
Pt
full-train-CNN 0.9037 0.9022 (± 0.0155)
CNN+SVM: 0.9646 0.9667 (± 0.0082)
fine-tune-CNN 0.9682 0.9678 (± 0.00092)
BIOIMAGING 2018 - 5th International Conference on Bioimaging
64
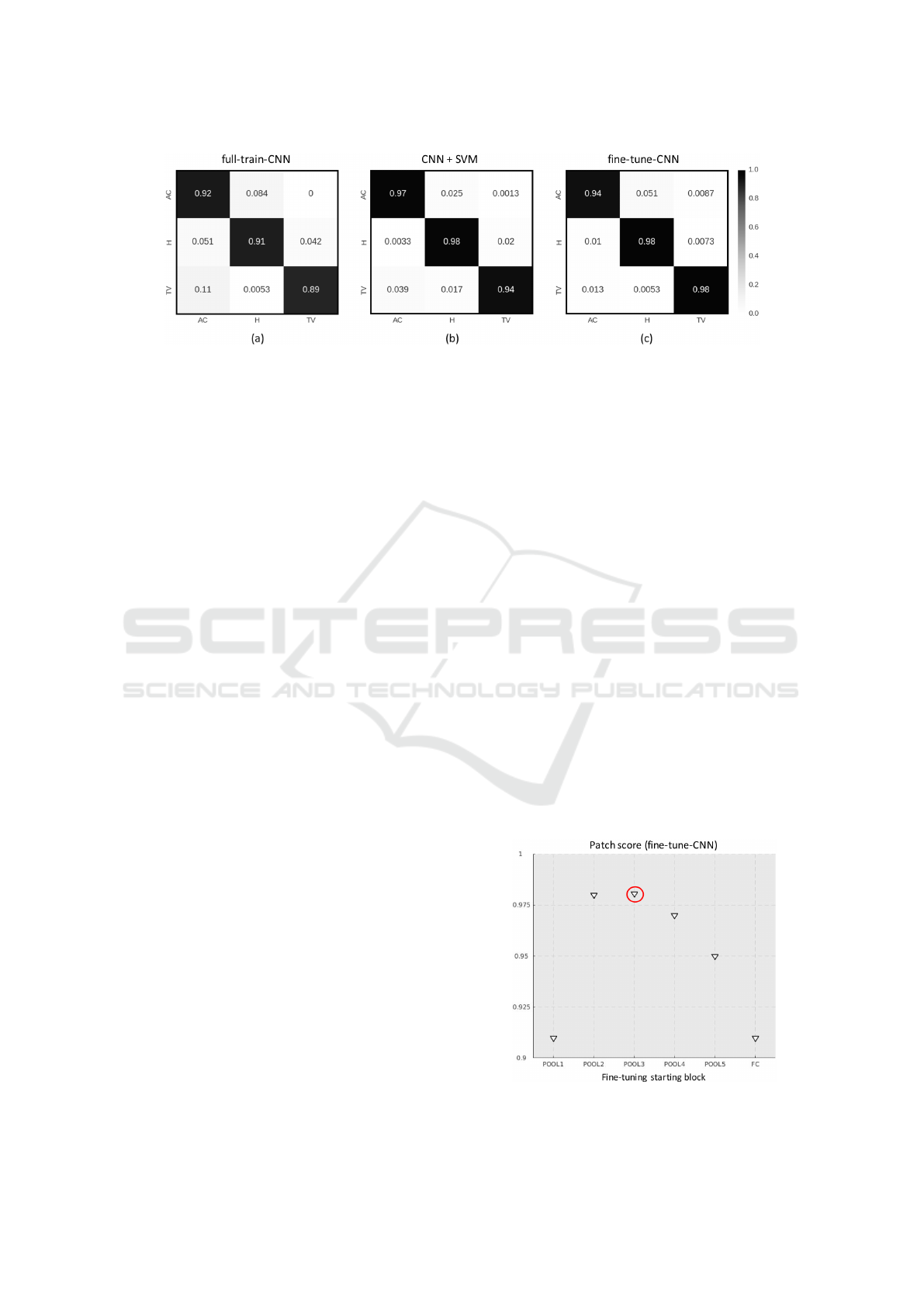
Figure 8: Patch-wise confusion matrices for (a) CNN fully trained on CRC samples, (b) SVM with pre-trained CNN as fixed
features generator, (c) pre-trained CNN with fine-tuning of the stages after POOL3 block.
the computational problems and overfitting risks
associated with full-training. Indeed, the fine-
tuning of the pre-trained CNN took only two
hours against the eight taken by full-training, us-
ing the same hardware and learning paradigm.
To investigate further on the performance of the
fine-tuned CNN, we run additional experiments by
changing the starting block for the backpropagation
algorithm. In Figure 9, we report the patch score ob-
tained on the test set, for different configurations of
the fine-tuning. In the x-axis, POOL-i means that only
the weights after the i-th POOL block were learnt on
the CRC training set, while all the rest of the parame-
ters were frozen to the values learnt on the ImageNet.
Likewise, FC means that only the fully-connected
stage of the network was trained. The trend of the
patch score values shows that the maximum accuracy
is reached when the CNN is fine-tuned after POOL3,
which confirms the qualitative results of t-SNE. On
top of that, we can observe that fully-training the net-
work obtains more or less the same results than train-
ing only the last fully-connected stage. This further
confirms that CNN can be successfully used to trans-
fer features learnt from the ImageNet.
4 CONCLUSIONS AND FUTURE
WORK
In this work we investigated the use of deep learning,
and more specifically of Convolutional Neural Net-
works, for the automated classification of colorectal
histology samples into three main classes of interest:
healthy tissue, adenocarcinoma or tubulovillous ade-
noma.
For this purpose, we applied a CNN with VGG16
architecture, which we fully trained on a large dataset
of pre-annotated images of colorectal samples. This
solution provided satisfactory results when applied to
an independent test dataset, with classification accu-
racy in the order of 90%.
Besides the traditional full training approach, we
investigated two types of transfer learning techniques:
(i) using the first convolutional stages of a CNN pre-
trained on the ImageNet as a fixed feature genera-
tor for a Support Vector Machine, with a preliminary
feature reduction step; (ii) using the colorectal train-
ing set to fine-tune the last convolutional and fully-
connected stages of the pre-trained CNN.
In our experiments the transfer learning tech-
niques outperform the full training approach both in
terms of classification accuracy (above 96%) as well
as in terms of training time. Hence, they demonstrate
that low-level features learnt by the CNN in a very dif-
ferent context (the ImageNet, in this case) can be suc-
cessfully transferred to the classification of colorectal
images.
As a future work, we plan to extend the classifi-
cation problem to more tissue categories (i.e. differ-
ent types of benign lesions, besides tubulovillous ade-
noma). In the long run, we plan to design a develop
a complete framework for the analysis of colorectal
WSIs based on CNNs.
Figure 9: Mean accuracy in relation to the first block till
back-propagation is continued.
Colorectal Cancer Classification using Deep Convolutional Networks - An Experimental Study
65

REFERENCES
A. Young, R. H. and Kerr, D. (2011). ABC of Colorectal
Cancer. Wiley-Blackwell, 2nd edition.
Chollet, F. et al. (2015). Keras.
https://github.com/fchollet/keras.
Di Cataldo, S. and Ficarra, E. (2017). Mining textu-
ral knowledge in biological images: Applications,
methods and trends. Computational and Structural
Biotechnology Journal, 15:56 – 67.
Hastie, T., Tibshirani, R., and Friedman, J. (2009).
Overview of supervised learning. In The elements of
statistical learning. Springer.
Janowczyk, A. and Madabhushi, A. (2016). Deep learn-
ing for digital pathology image analysis: A compre-
hensive tutorial with selected use cases. Journal of
Pathology Informatics, 7(1):29.
Korbar, B., Olofson, A. M., Miraflor, A. P., Nicka, C. M.,
Suriawinata, M. A., Torresani, L., Suriawinata, A. A.,
and Hassanpour, S. (2017). Deep learning for clas-
sification of colorectal polyps on whole-slide images.
Journal of Pathology Informatics, 8:30.
Maaten, L. v. d. and Hinton, G. (2008). Visualizing data
using t-sne. Journal of Machine Learning Research,
9(Nov):2579–2605.
Marley, A. R. and Nan, H. (2016). Epidemiology of col-
orectal cancer. International Journal of Molecular
Epidemiology and Genetics, 7(3):105–114.
Qian, N. (1999). On the momentum term in gradi-
ent descent learning algorithms. Neural networks,
12(1):145–151.
Simonyan, K. and Zisserman, A. (2014). Very deep con-
volutional networks for large-scale image recognition.
arXiv preprint arXiv:1409.1556.
Weiss, K., Khoshgoftaar, T. M., and Wang, D. (2016). A
survey of transfer learning. Journal of Big Data,
3(1):9.
Yao, Y., Rosasco, L., and Caponnetto, A. (2007). On early
stopping in gradient descent learning. Constructive
Approximation, 26(2):289–315.
BIOIMAGING 2018 - 5th International Conference on Bioimaging
66
