
Robust Plant Segmentation from Challenging Background with a
Multiband Acquisition and a Supervised Machine Learning
Algorithm
Taha Jerbi, Aaron Velez Ramirez and Dominique Van Der Straeten
Laboratory of Functional Plant Biology, Dept. of Biology, Ghent University, K. L. Ledeganckstraat 35, 9000 Gent, Belgium
Keywords: Image Processing, Plant Phenotyping, Classification, Segmentation, Multispectral Acquisition, Supervised
Learning, Arabidopsis.
Abstract: Remote sensing through imaging forms the basis for non-invasive plant phenotyping and has numerous
applications in fundamental plant science as well as in agriculture. Plant segmentation is a challenging task
especially when the image background reveals difficulties such as the presence of algae and moss or, more
generally when the background contains a large colour variability. In this work, we present a method based
on the use of multiband images to construct a machine learning model that separates between the plant and
its background containing soil and algae/moss. Our experiment shows that we succeed to separate plant parts
from the image background, as desired. The method presents improvements as compared to previous methods
proposed in the literature especially with data containing a complex background.
1 INTRODUCTION
Accurate segmentation of plants from the image
background is the first step for the extraction of traits
in phenotyping applications and precision agriculture.
An intuitive approach that simplifies the task by
covering the soil (Arend et al., 2016) could be the
easiest solution making the segmentation
straightforward; however, this solution is not
practical in field applications. Moreover, it could
perturb the gas exchange between the soil and the air
in laboratory experiments. Image processing based on
algorithms which succeed to overcome application-
specific difficulties offers solutions to extract plant
traits. Image segmentation difficulties include: (a)
large variability of the soil colour, (b) the presence of
moss and algae with colours similar to the plant to
segment, (c) the presence of non-green plant parts as
flowers and/or yellow and brown parts of leaves of
senescing plants or in stressed plants, which can
occur in phenotyping experiments under specific
conditions.
Previous work tackled this problem mainly by
considering a classification problem on the colour
space. Sharr (Sharr et al., 2016) presents four
segmentation methods from different universities and
research laboratories (Leibniz Institute of Plant
Genetics and Crop Plant Research-IPK-Germany,
Nottingham University-United Kingdom, Michigan
State University-United States, and Wageningen
University-Netherlands) in a challenge frame. The
first method uses a 3D histogram cube to encode the
probability for each observed pixel in the training
data of belonging to the plant or background. The
second method is based on a superpixel segmentation
in Lab colour space as a first step, and thresholding of
the superpixel image in the second step. In the third
method, the foreground/background segmentation is
done by a simple empirical threshold on the ‘a’
channel of the Lab colour space. The fourth method
uses an artificial neural network (ANN) with one
hidden layer for plant/background separation. In
general, for all these methods and as mentioned in the
paper (Sharr et al., 2016), the results show that “most
methods perform well in separating plant from
background, except when the background presents
challenges” (Sharr et al., 2016). A similar approach
using the watershed algorithm was also proposed in
an earlier study for plant segmentation from
background (Åstrand et al., 2006).
In (De Vylder et al., 2012), the authors used the
Expectation Maximization (EM) algorithm with the
hue signal of the images to characterize two
Gaussians distributions modeling the plant and the
Jerbi T., Velez Ramirez A. and Van Der Straeten D.
Robust Plant Segmentation from Challenging Background with a Multiband Acquisition and a Supervised Machine Learning Algorithm.
DOI: 10.5220/0006552001000105
Copyright
c
2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

Figure 1: System acquisition of Arabidopsis plants.
background in the histogram. However, this method
will also face difficulties when the hue signal of the
plant and the background are similar.
More recently, machine learning approaches have
been used to segment plants from soil (Navarro et al.,
2016), but the images used were the Red-Green-Blue
(RGB) and the Near Infra-Red (NIR) and the
segmentation was done separately for the RGB and
NIR. In this study, a comparison between three
machine learning approaches (k-nearest neighbor
(kNN), naive Bayes classifier (NBC), Support Vector
Machine (SVM)) showed that SVM performed better
for the NIR, while kNN segmentation was better for
the colour images (RGB).
The majority of algorithms cited above use either
one channel or the three colour channels RGB except
the fourth algorithm cited in (Sharr et al., 2016) that
uses also an excessive green value (R,2G,B) with two
texture features, and the machine learning approach
in (Navarro et al., 2016) that uses the NIR signal.
However, in phenotyping applications, more signals
are generally acquired to observe the plant response
to different wavelengths including fluorescence
signals. In addition to their biological meaning, these
data can also be useful for the plant segmentation. In
our project, which is part of the TIMESCALE
project-Horizon 2020, 14 bands are used to observe
the plants’ responses to different environmental
conditions. These bands are used in our segmentation
approach as discussed below. In the next section, we
present our acquisitions and the algorithm proposed
for a better segmentation of the plant from a
challenging background. In section 3, we evaluate our
results and discuss the different methods.
2 MATERIALS AND METHODS
2.1 Materials and Data
Our phenotyping platform is composed by a table, a
robot, an acquisition system (lighting system, filters
and cameras) and software that automatically
acquires images during a biological experiment. The
acquisition system is composed by cameras and filters
that allow acquisition at specific wavelengths.
Chlorophyll fluorescence signals allow to study plant
photosynthesis (Baker, 2008). The system flashes a
specific wavelength in a very short interval of time
and captures the emitted signal by the plant
photosynthetic apparatus. Our system allows to
measure these signals in different phases: an excited
phase when the plant is under light and a non-excited
phase when the plant is in darkness. Different bands
are either directly acquired by the camera and the
filter system or computed from the acquired bands.
These signals and their biological interpretations are
described in (Baker, 2008) and in (Gitelson et al.,
1999). For each acquisition, the robot moves the
acquisitions system (cameras and filters) on top of a
plate containing plants and acquires images. All
bands can be acquired in less than 2 minutes, and we
assume there is no modification of the scene during
this short period of time (no shape or position
variation).
Figure 2: Acquisition of an Arabidopsis plant with 3 different bands of the system, the differentiation between the plant and
the algae/moss is difficult with each wavelength band shown.

(a) (b) (c) (d)
Figure 3: Plant segmentation in presence of moss and algae: (a) Plant from our database, (b) Our automatic segmentation
(RF), (c) Plant from the database of (Scharr, 2016), (d) Our automatic segmentation (SVM Gaussian).
In our experiment, plants were imaged from day 5
after sowing (Das 5) to Das 22. For the purpose of the
biological experiment, some plants were under
drought stress. The system generated approximate-
vely 30000 acquisitions (14 images each) for an
experiment that takes 22 days. Each image contains
36 plants.
2.2 Method
First, as a pre-processing step, the images were split
into smaller images containing one plant each. We
considered 115 images to form our database. If
needed, images were downsized to have pixel to pixel
correspondence between all bands. In addition to the
14 bands registered, 6 more bands were computed:
the HSV (3 bands that represent the colour images as
Hue, Saturation and Value), and to include informa-
tion about the neighbors of a pixel, we also computed
the median value in a 15 pixels large square (H15,
S15 and V15) using a median filter. Altogether, 20
features are available to characterize the plants and
the background (soil, moss and algae) pixels in the
image.
In our approach, pixel classification is used as a
method for plant segmentation. To overcome the
difficulties of the plant separation from a complex
background, supervised machine learning approaches
bring a solution to have a specific model to a
particular dataset. In fact, if the dataset contains
plants with flowers, the model constructed will be
different from a model constructed from plants with
just leaves for example. Two main supervised
machine learning approaches were used in this work,
the support vector machine algorithm (SVM) and the
random forest algorithm (RF).
SVM is a supervised machine learning approach
that was introduced by Vapnik and Cortes (Cortes and
Vapnik, 1995). The method is based on two steps. In
the first one (learning or training step), based on pre
labelled samples, the algorithm constructs borders to
separate data into classes (regions) defined by the
predefined labels (figure 4.). It also ensures maximum
margins between the borders and the samples in order
to reduce errors when applying these borders to new
data. In the second step (prediction step), the model
(borders) is applied to the new data to predict the
classes of the new input.
Figure 4: Training data representation in 3D: plant data in
green and background data in red (only RGB colour
features are represented). Dataset: Ara2013 (Scharr et al.,
2016), Number of images: 10/165.
Random Forest (RF) algorithm (Breiman, 2001)
is also a supervised algorithm. It is based on the idea
of using the decision trees where each tree is
constructed using bootstrapped random variables. In
each tree node, the decision is taken according to the
best among random predictors. By generating
multiple trees, the random forest algorithm is
obtained. At the end of the algorithm, the forest (all
the trees) takes a decision corresponding to the global
vote of its trees.
In order to construct our models with both
methods, the pixel values are taken as features in
order to use not only colours but also the fluorescent
responses of the plants. In addition, to consider the
presence of noise and to integrate an information
about the neighbourhood of a pixel for the decision of
its class, we include in the features the outputs of the
median filter (size 15) on the channels H, S and V, so
that a pixel having values corresponding to a plant but
with neighbourhood values corresponding to the
background, will not be considered automatically as
a plant.
In all, we obtain 20 features that could be used
with our models and the problem can be stated as
looking for a classifier that allows predicting the
labels from the available features (see eq 1). 11
images were selected from our database to construct
the models.
Given a set of labeled samples ,
(1)
Look for that verifies:
with is the input vector, is the number of bands,
is the label and is the model to construct
(the classifier).
Using the SVM and the RF algorithms, we
constructed different models in order to determine the
best segmentation method for our application. Using
the SVM, we built 3 models: the first one uses only 6
features: H, S, V and H15, S15, V15 and has Gaussian
kernels, the second uses the same 6 features with
linear kernels and the third one uses all features (20)
with linear kernels. The construction of an SVM
model using all bands (20) and Gaussian kernels
would generate time computation issues and was
avoided in this work for this reason. With the RF
algorithm, we constructed 2 models, the first one uses
the 6 features previously cited (H, S, V and H15, S15,
V15) and the second uses all features (20). The
number of trees is 200 for both RF models. All in all,
we have 5 models: 3 SVM models and 2 RF models.
We also can distinguish our models as, 3 models
using just 6 features and 2 models using all available
features (20). Table 1, shows the evaluation of the
results obtained with these different models in
comparison to the ground truth segmentation made
manually by an expert as it will be explained in the
next section.
3 RESULTS AND DISCUSSION
To compare our method to the state of the art, we used
the dataset Ara2013 containing 165 images and
published in (Sharr et al., 2016). Since the database
contains only RGB images, we trained our model
with just 6 features (3 colours and 3 neighbors mean)
for this test. Ten images where considered to
construct the model. The sensitivity, the accuracy and
the precision of our segmentation method using the
SVM model with the Gaussian kernel are shown in
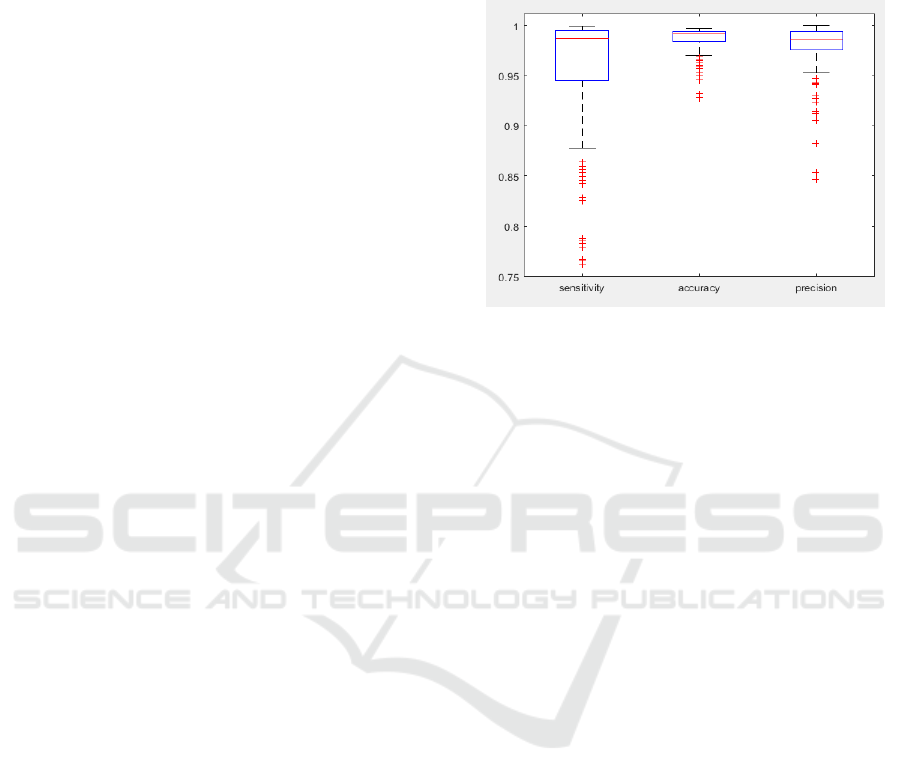
figure 5.
Figure 5: Performance of the method with the set of images
described in (Sharr et al, 2016). Sensitivity, accuracy and
precision are used for the evaluation.
To make a comparison with the previous methods
results, we used the Foreground-Background distance
which measures the difference between the sum of the
difference in the automatic and ground truth
segmentation as mentioned and used in (Sharr et al.,
2016) (see eq 2), we obtained a result of 96,8 (3,4)
which is similar to best reported results with the IPK-
Germany algorithm 96.3 (1.7). This result was
expected since the dataset does not contain a large
amount of images with algae and moss. Other
methods (Nottingham-UK, MSU-USA, Wageningen-
Netherlands) reported in the paper provided
respectively these results: 93.0 (4.2); 87.7 (3.6) and
95.1 (2.0).
(2)
with is the foreground-background distance,
is the automatic segmentation and
is the ground
truth segmentation.
In a second step of validation, we asked an
independent expert to manually segment the plants in
our images. The manual segmentation was done using
the RGB channels and the segmentation of the other
channels were obtained by applying the mask from
the segmented one. An evaluation of the automatic
segmentation was done by computing the sensitivity,
the accuracy and the precision as shown in the
equation 3:
(3)
with TP, the true positive, TN true negative, FP the
false positive and FN the false negative values of the
data. This evaluation system measures the proportion
of positive pixels that are correctly classified
(sensitivity), gives an indication of the uniformity and
the reproducibility of the classification (precision)
and evaluates the proportion of the true results in
comparison to the reference classification (accuracy).
This evaluation system allows a better appreciation of
the classification than the Foreground-Background
distance (see eq. 2) used in (Sharr et al., 2016) as the
latter fuses the errors in the background and in the
plants (foreground).
Table 1: SVM and RF segmentations evaluation: mean
values of sensitivity, accuracy and precision based on the
comparison between the automatic segmentation and the
expert segmentation.
Sensitivity
Accuracy
Precision
SVM 6
Bands linear
0.754903
0.959994
0.889562
SVM 6
Bands Gauss
0.870557
0.984172
0.994886
SVM 20
Bands linear
0.830052
0.982195
0.985209
RF 6
Bands
0.898015
0.986163
0.987321
RF 20
Bands
0.900721
0.987252
0.993303
To evaluate the errors generated by manual
segmentation, an intra-user segmentation evaluation
was made. First, one manual image segmentation was
taken as a gold standard. Then, 3 different
segmentations of the same image were performed by
the same user. The results showed a very small
variability due to manual segmentation (sensitivity
0.989426; precision 0.993021; accuracy 0.992546).
For this reason, we can consider, with a high
confidence, the expert manual segmentation of our
data as a gold standard and a reference to the
automatic approach.
Globally, the constructed models gave good
results, as shown in table 1 and figure 3. In the SVM
models, the Gaussian kernels gave a better results even
using just 6 bands, than the linear kernels. This can be
explained by the fact that the problem is not linear. The
reduction of the computation time by choosing linear
kernels results in poorer sensitivity and even using all
the bands, the results with linear kernels do not
outperform the Gaussian kernels with just 6 bands.
However, the use of all bands with linear SVM kernel
makes notable increase of the segmentation
performance when comparing to the same approach
(SVM linear kernels) using just 6 bands (sensitivity
varies from 0.7549 to 0.83; accuracy 0.959 to 0.982
and precision 0.889 to 0.985).
Comparing the SVMs and the RF approaches,
both RF methods gave better results than the SVM
methods, especially for the sensitivity that increases
from 0.87 (best SVM: SVM with Gaussian kernel) to
0.898 and 0.9. The use of all the 20 features increases
slightly the sensitivity and the precision in the RF
approach while this improvement is more
considerable with the linear SVM approach as shown
in table 1.
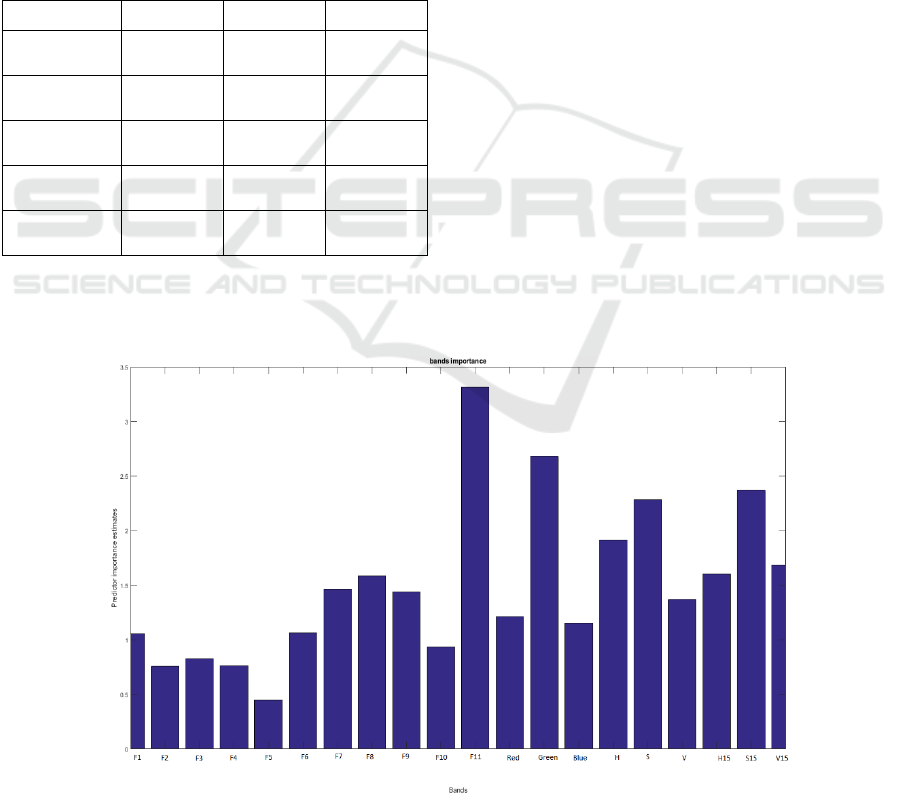
Figure 6: Bands contribution into the Random Forest segmentation.

In figure 6, the contribution of each band is
evaluated in the construction of the RF model with all
bands. The Green G, the Saturation S, and the
Saturation neighbours S15 are among those who
contribute the most but the highest contribution is
made by the F11 fluorescence band. This band
expresses the chlorophyll content of the plant as
described in (Gitelson et al., 1999), which explains
the high contribution of this band in the classification
of plant versus soil with no chlorophyll and algae and
moss with different chlorophyll composition. The
bands H15, S15 and V15 are also highly contributing
to the RF models constructions. These bands could
also be interpreted as noise reduced HSV signals, and
we can notice that the contribution of S15 was even
bigger than the contribution of S which indicates the
importance of its use.
4 CONCLUSIONS
As a conclusion, both used supervised machine
learning algorithms: SVM and RF approaches
succeeded to provide a useful tool for the plant
segmentation in the presence of challenging
background containing algae and moss. RF
approaches gave better results than the SVM
methods. The use of multiple bands showed that the
performance of the algorithms improves the
segmentation results especially with the SVM
models. Within the RF approach, bands contribution
to the final results vary and the highest contribution is
the ratio fluorescence which highlights the role of
these bands in such machine learning approaches. In
addition, the neighbourhood information introduced
by the channels H15, S15, and V15 contributed
considerably to the construction of the model and to
the improvement of the segmentation results.
One limitation of this approach is that it is based
on the expert knowledge constructing the data needed
for training the model (supervised machine learning).
In this sense, bad quality labelled image (a bad expert
segmentation) will result in a bad model for
segmentation.
To avoid this dependence, we will develop, in
future work, unsupervised models for the segmenta-
tion of the plants. Moreover, more features will be
included in the classification parameters such as
texture features to improve the segmentation results.
We will also focus on the plant 3D acquisition to
obtain more data (such as height and volume)
describing its responses to the environment (drought,
nutrients, biotic stress). These 3D information will
also be helpful for the segmentation and the time
tracking of individual plant leaves.
ACKNOWLEDGEMENTS
The authors thank Joke Belza for her help with the
manual segmentation of the data.
This work was financed by TIMESCALE, a
HORIZON 2020 project, and Ghent University.
REFERENCES
Arend D., Lange M., Pape J-M., Weigelt-Fischer K.,
Fernando Arana-Ceballos, Mücke I., Klukas C.,
Altmann T., Scholz U., and Junker A. 2016, “Data
Descriptor: Quantitative monitoring of Arabidopsis
thaliana growth and development using high-
throughput plant phenotyping”, Scientific Data 3,
Article number: 160055.
Åstrand B., and Johansson M. 2006; Segmentation of
partially occluded plant leaves; 13th International
Conference on Systems, Signals and Image Processing;
September 21-23, 2006, Budapest, Hungary.
Baker N. R. 2008, Chlorophyll Fluorescence: A Probe of
Photosynthesis In Vivo, Annual Review of Plant
Biology, 59:89-113.
Breiman L. 2001, “Random Forests”, Machine Learning,
pp 5–32, October 2001, Volume 45, Issue 1.
Cortes C., Vapnik V. 1995, “Support-Vector Networks”,
Machine Learning, 20, 273-297.
Gitelson A. A., Buschmann, C., and Lichtenthaler H. K.
1999; The Chlorophyll Fluorescence Ratio F735/F700
as an Accurate Measure of the Chlorophyll Content in
Plants, Remote Sensing Environment 69:296–302.
Navarro P. J., Pérez F., Weiss J., and Egea-Cortines M.
2016, “Machine Learning and Computer Vision System
for Phenotype Data Acquisition and Analysis in
Plants”, Sensors (Basel). 2016 May; 16(5): 641.
Scharr H., Minervini M., French A. P., Klukas Ch., Kramer
D. M., Liu X., Luengo I., Pape J-M., Polder G.,
Vukadinovic D., Yin X., and Tsaftaris S. A. 2016, “Leaf
segmentation in plant phenotyping: a collation study”,
Machine Vision and Applications 27:585–606.
De Vylder J., Vandenbussche F., Hu Y., Philips W., and
Van Der Straeten D. 2012, “Rosette Tracker: An Open
Source Image Analysis Tool for Automatic
Quantification of Genotype Effects.”, Plant Physiology,
November 2012, Vol. 160, pp. 1149–1159.
