
On the Application of Bio-Inspired Optimization Algorithms to Fuzzy
C-Means Clustering of Time Series
Muhammad Marwan Muhammad Fuad
Forskningsparken 3, Institutt for kjemi, NorStruct
The University of Tromsø - The Arctic University of Norway, NO-9037 Tromsø, Norway
Keywords: Data Mining, Differential Evolution, Distance Metrics, Fuzzy C-Means Clustering, Genetic Algorithms,
Overfitting, Particle Swarm Optimization, Time Series.
Abstract: Fuzzy c-means clustering (FCM) is a clustering method which is based on the partial membership concept.
As with the other clustering methods, FCM applies a distance to cluster the data. While the Euclidean
distance is widely-used to perform the clustering task, other distances have been suggested in the literature.
In this paper we study the use of a weighted combination of metrics in FCM clustering of time series where
the weights in the combination are the outcome of an optimization process using differential evolution,
genetic algorithms, and particle swarm optimization as optimizers. We show how the overfitting
phenomenon interferes in the optimization process that the optimal results obtained during the training stage
degrade during the testing stage as a result of overfitting.
1 INTRODUCTION
A time series is an ordered collection of
observations at intervals of time points. These
observations are real-valued measurements of a
particular phenomenon.
Time series arise in disciplines ranging from
medicine, and finance to meteorology, and
engineering, to name a few. For this reason, time
series data mining has received increasing attention
over the last two decades.
Time series data mining handles several tasks
such as classification, clustering, similarity search,
motif discovery, anomaly detection, and others. All
these tasks are based on the concept of similarity
which is measured using a similarity measure or a
distance metric.
Bio-inspired Optimization, also called nature-
inspired Optimization, is a branch of optimization
which, as the name suggests, is inspired by natural
phenomena. It has diverse applications in
engineering, finance, economics and other domains.
In computer science bio-inspired optimization has
been applied in several fields such as networking,
data mining, and software engineering.
As with other fields of computer science,
different papers have proposed applying bio-inspired
optimization to data mining tasks (Muhammad Fuad,
2012a, 2012b, 2014a).
In this work we study the application of three
well-known bio-inspired optimization algorithms:
differential evolution, genetic algorithms, and
particle swarm optimization, to the task of FCM
clustering of time series where these optimizers are
used to obtain the optimal values of the weights
assigned to the distances that constitute a
combination of metrics used in the FCM clustering
task.
The rest of the paper is organized as follows; in
Section 2 we present background on time series,
clustering, similarity measures, and overfitting. In
Section 3 we introduce the proposed method which
we test in Section 4. The results of the experiments
are discussed in Section 5. We conclude this paper
with Section 6.
2 BACKGROUND
A time series is an ordered collection of observed
data. Formally, a time series S of length n is defined
as;
nnn
tvstvstvsS ,,....,,,,
222111
.
These values can be real numbers or multi-
dimensional vectors.
348
Muhammad Fuad M..
On the Application of Bio-Inspired Optimization Algorithms to Fuzzy C-Means Clustering of Time Series.
DOI: 10.5220/0005276203480353
In Proceedings of the International Conference on Pattern Recognition Applications and Methods (ICPRAM-2015), pages 348-353
ISBN: 978-989-758-076-5
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

Data mining is the analysis of data with the goal
of uncovering hidden knowledge in it. Data mining
includes several tasks such as classification,
clustering, anomaly detection, and others.
Processing these different tasks usually requires
extensive computing.
Clustering is a fundamental data mining task.
Clustering seeks for the grouping tendencies of the
data. Clustering can be defined as the process of
finding natural groups, called clusters, in a dataset.
The objective of the clustering task is to find the
most homogeneous clusters that are as distinct as
possible from other clusters. This grouping should
maximize inter-cluster variance while minimizing
intra-cluster variance (Esling and Agon, 2012).
There are several basic clustering methods, those
which concern time series data are Partition-based
Clustering, Hierarchical-based Clustering, and
Model-based Clustering (Liao, 2005). Figure 1
shows the general scheme of the partition-based
clustering algorithm.
Fuzzy c-means Clustering (FCM), is a partition-
based clustering method. FCM is based on the
concept of Partial Membership where the allocation
of the data point to the centers becomes a matter of
degree – the higher the membership value, the
stronger its bond to the cluster (Krzysztof et al,
2007).
The objective of FCM clustering is to minimize
the measure:
ip
c
p
n
i
m
pim
xzduE ,
2
11
(1)
Where u is the fuzzy membership and m is fuzzy
exponent. In the following we present a brief
description of FCM (Maulik et al, 2011). FCM starts
with random c initial cluster centers. In the
following step, and at every iteration, the algorithm
finds the fuzzy membership of each data object to
every cluster using the following equation:
1
2
1
,
,
1
mc
j
ij
ip
pi
xzd
xzd
u
(2)
for 1 ≤ p ≤ c, 1 ≤ i ≤ n.
Al
g
orithm Clustering
Require c
t
he number of clusters,
n data objects.
1.
(
Randomly) initialize the
cluster centers.
2.
F
or each data object, and
for each center, comput
e
t
he distance between this
d
ata object and the cluster
c
enter, assign the data
o
bject to the closest
center.
3. Update the cluster centers.
4. Repeat steps 2-3 until
convergence.
Figure 1: The partition-based clustering algorithm.
Based on the membership values, the cluster centers
are recomputed using the following equation:
n
i
m
pi
i
n
i
m
pi
p
u
xu
z
1
1
(3)
1 ≤ p ≤ c
The algorithm continues until a stopping condition
terminates it.
2.1 Overfitting
One of the frequent problems encountered when
generating a data mining model is overfitting.
Overfitting results when the provisional model tries
to account for every possible trend or structure in the
training set. In overfitting the algorithm memorizes
the training set at the expense of generalizability to
the validation set (Larose, 2005). There are several
reasons for overfitting, and overfitting-avoidance is
an important research topic in data mining. The
question of overfitting is particularly important
when applying bio-inspired algorithms to perform
data mining tasks (Muhammad Fuad, 2014b).
2.2 Similarity Measures
Measuring the similarity between two time series, on
which clustering and other data mining tasks are
based, is not a trivial task. There have been several
distance metrics and similarity measures proposed in
OntheApplicationofBio-InspiredOptimizationAlgorithmstoFuzzyC-MeansClusteringofTimeSeries
349

the literature. The most common distance metric,
however, is the Minkowski distance, which is
defined between two time series S and T as:
p
n
i
p
iip
tsTSL
1
),(
(4)
If p =1, the distance is called the Manhattan distance
or the city block distance. If p = ∞, it is called the
infinity distance or the chessboard distance, and if p
= 2 we get the widely-known Euclidean distance.
Applying any of these distances takes linear time in
the length of the series.
3 APPLYING A COMBINATION
OF DISTANCES TO FCM
CLUSTERING
The idea of applying a combination of distance
metrics or similarity measures to handle data mining
tasks has been proposed by different researchers. In
(Bustos and Skopal, 2006) the authors propose using
a weighted combination of several metrics to handle
the similarity search problem. The novelty of our
work is that we address this as an optimization
problem. More formally, we FCM cluster the time
using a weighted combination of distance metrics
defined as:
n
1i
ii
T,SdT,Sd
(5)
where
1,0
i
.
In order to perform this optimization process,
and for comparison reasons, we choose to use three
of the most powerful bio-inspired optimization
algorithms which are Differential Evolution, Particle
Swam Optimization, and Genetic Algorithms.
3.1 Differential Evolution
Differential Evolution (DE) is an evolutionary
optimization algorithm which is particularly adapted
to solve continuous optimization problems.
DE starts with a population of PopSize vectors
each of which is of nbp dimensions, where nbp is the
number of parameters (in our problem, nbp is the
number of distances in the combination given by
equation (5)). In the next step for each individual
i
T
(called the target vector) of the population three
mutually distinct individuals
1r
V
,
2r
V
,
3r
V
and
different from
i
T
are chosen randomly from the
population. The donor vector
D
is formed as a
weighted difference of two of
1r
V
,
2r
V
,
3r
V
, added
to the third; i.e.
3r2r1r
VVFVD
.
F
is called
the mutation factor.
The trial vector
R
is formed from elements of
the target vector
i
T
and elements of the donor
vector
D
according to different schemes. In this
paper we choose the crossover scheme presented in
(Feoktistov, 2006). In this scheme an integer
Rnd is
chosen randomly among the dimensions
nbp,1 .
Then the trial vector
R
is formed as follows:
otherwiset
iRndC,randifttFt
t
j,i
rj,ir,ir,ir,i
i
10
321
(6)
where
nbp,...,i 1
.
r
C is the crossover constant. In
the next step DE selects which of the trial vector and
the target vector will survive in the next generation
and which will die out. This selection is based on
which of
i
T
and R
yields a better value of the fitness
function. DE continues for a number of generations
NrGen.
3.2 The Genetic Algorithm
The Genetic Algorithm (GA) is another member of
the evolutionary algorithms family. GA has the
following elements: a population of individuals,
selection according to fitness, crossover to produce
new offspring, and random mutation of the new
offspring (Mitchell, 1996).
The first step of GA is defining the problem
variables and the fitness function. A particular
configuration of variables produces a certain value
of the fitness function and the objective of GA is to
find the values of the parameters that optimize the
fitness function. As with DE, GA starts with a
population of
PopSize vectors (also called
chromosomes) each of which is of nbp dimensions.
Each chromosome represents a possible solution to
the problem at hand. The fitness function of each
chromosome is evaluated. The next step is
selection.
The purpose of this process is to determine which
chromosomes are fit enough to survive and possibly
produce offspring and which will die out. This is
decided according to the fitness function of each
chromosome. The percentage of chromosomes
selected for mating is denoted by
sRate. Crossover is
the next step in which offspring of two parents are
produced to enrich the population with fitter
ICPRAM2015-InternationalConferenceonPatternRecognitionApplicationsandMethods
350

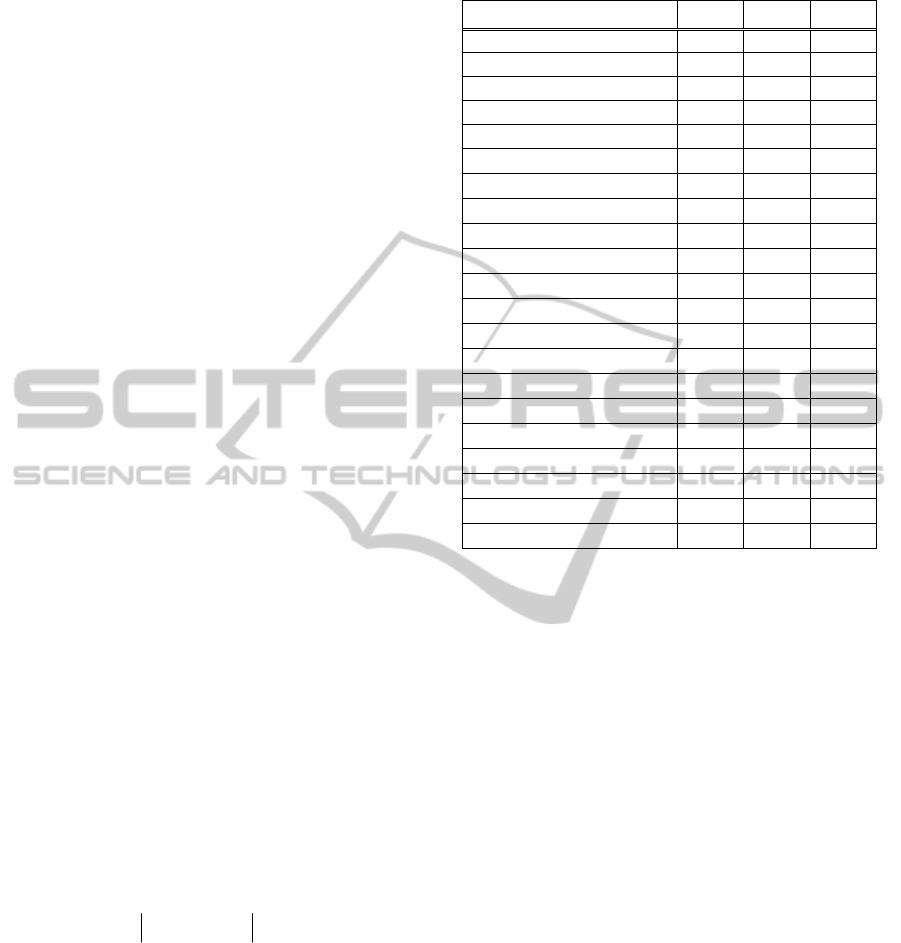
Table 1: The FCM clustering errors on the training sets and testing sets for DE, PSO, and GA, together with the FCM
clustering errors for L
1
, L
2
, L
∞
.
Dataset L
1
L
2
L
∞
DE PSO GA
E
train
E
test
E
train
E
test
E
train
E
test
Beef 0.49 0.49 0.45 0.36 0.48 0.36 0.46 0.36 0.49
CBF 0.48 0.50 0.51 0.30 0.51 0.30 0.51 0.30 0.49
CinC_ECG_torso 0.75 0.77 0.77 0.42 0.69 0.51 0.75 0.51 0.76
Coffee 0.50 0.50 0.46 0.36 0.51 0.36 0.50 0.36 0.50
ECG200 0.30 0.28 0.25 0.22 0.28 0.26 0.30 0.25 0.26
ECGFiveDays 0.50 0.48 0.46 0.45 0.50 0.45 0.49 0.45 0.50
FaceFour 0.43 0.57 0.68 0.24 0.56 0.26 0.57 0.29 0.46
FISH 0.67 0.64 0.69 0.56 0.62 0.57 0.63 0.57 0.63
Gun_Point 0.47 0.48 0.46 0.36 0.47 0.36 0.47 0.38 0.48
Lighting7 0.67 0.82 0.67 0.54 0.64 0.60 0.70 0.58 0.66
MedicalImages 0.84 0.79 0.72 0.70 0.82 0.72 0.82 0.72 0.79
MoteStrain 0.17 0.16 0.27 0.10 0.17 0.15 0.15 0.15 0.17
OliveOil 0.52 0.33 0.30 0.14 0.46 0.22 0.39 0.25 0.36
OSULeaf 0.80 0.82 0.86 0.71 0.79 0.74 0.83 0.74 0.80
SonyAIBORobotSurfaceII 0.23 0.22 0.23 0.15 0.23 0.15 0.23 0.15 0.23
SwedishLeaf 0.77 0.80 0.88 0.69 0.71 0.70 0.81 0.70 0.80
Symbols 0.38 0.39 0.29 0.09 0.40 0.14 0.40 0.14 0.38
synthetic_control 0.54 0.53 0.47 0.35 0.57 0.39 0.54 0.42 0.51
Trace 0.48 0.48 0.43 0.38 0.48 0.39 0.45 0.39 0.46
TwoLeadECG 0.47 0.47 0.47 0.31 0.47 0.35 0.49 0.35 0.47
yoga 0.48 0.49 0.48 0.43 0.48 0.43 0.48 0.48 0.48
chromosomes. GA also applies a mechanism called
mutation through which a percentage mRate of
genes is randomly altered. The above steps repeat
for
NrGen generations.
3.3 Particle Swarm Optimization
Particle Swarm Optimization (PSO) is a member of
a family of naturally-inspired optimization
algorithms called
Swarm Intelligence (SI). PSO is
inspired by the social behavior of some animals,
such as bird flocking or fish schooling (Haupt and
Haupt, 2004). In PSO individuals, called
particles,
follow three rules a)
Separation: each particle avoids
getting too close to its neighbors. b) Alignment: each
particle steers towards the general heading of its
neighbors, and c)
Cohesion: each particle moves
towards the average position of its neighbors.
PSO starts by initializing a swarm of
PopSize
particles at random positions
0
i
X
and velocities
0
i
V
where
PopSizei ,..,1
. In the next step the
fitness function of each position, and for each
iteration, is evaluated. The positions
1k
i
X
and
velocities
1k
i
V
are updated at time step ( 1
k )
according to the following formulae:
k
i
k
iL
k
i
k
G
k
i
k
i
XLXGV.V
1
k
i
k
i
k
i
VXX
1
(8)
where
GGG
ar .
,
LLL
ar .
,
1,0, Urr
LG
,
R
GL
aa ,,
,
k
i
L
is the best position found by
particle i,
k
G
is the global best position found by the
whole swarm,
is called the inertia ,
L
a is called
the local acceleration , and
G
a is called the global
acceleration. The algorithm continues for a number
of iterations.
4 EXPERIMENTS
The objective of our experiments is to see if a
weighted combination of the Euclidean distance, the
Manhattan distance, and the infinity distance will
give better FCM clustering results than when each of
these distances is used as a stand-alone distance.
More formally, we FCM cluster the data using the
following combination:
T,SLT,SLT,SLT,Sd
32211
(9)
As indicated earlier the weights
i
are obtained
OntheApplicationofBio-InspiredOptimizationAlgorithmstoFuzzyC-MeansClusteringofTimeSeries
351
through an optimization process that minimizes the
FCM cluttering error as given by equation (1).
We tested our method on a variety of time series
datasets from the UCR archive (Keogh et al, 2011).
We meant to choose datasets of different sizes and
lengths to get unbiased results.
In our experiments we compared the three
optimizers presented in Section 3 in terms of FCM
clustering error.
We used the same control parameters for the
three optimizers (except for those which are proper
to that certain optimizer). PopSize was set to 16 and
NrGen was set to 100.
For each dataset, and for each optimizer, the
experiment consists of two stages; the training stage
and the testing stage. In the training stage we
perform an optimization process where the
parameters of the optimization problem are the
weights
3,2,1; i
i
. The outcome of this
optimization problem is the weights ω
i
which yield
the minimum FCM clustering error. These optimal
values are then applied to the testing sets. In Table 1
we present the results of our experiments.
Taking into account that the optimization process
was applied to the training datasets, we see that the
three optimizers did improve the FCM clustering as
the FCM clustering errors of the training datasets for
all the datasets, and whatever optimizer is used, are
smaller than the errors resulting from using L
1
, L
2
,
or L
∞
, as stand-alone distances. However, when
comparing the FCM clustering errors on the testing
datasets we see that these errors are larger than those
obtained when using L
1
, L
2
, or L
∞
, as stand-alone
distances.
In order to understand this phenomenon, we
investigate whether the learning process has
undergone overfitting.
There are several measures in the literature to
measure overfitting. We opted for the following
simple measure:
testtrain
EEOF
(10)
Where E
train
,, E
test
, are the FCM clustering errors on
the training set and the testing set, respectively, OF
is the overfitting value.
In Table 2 we report the overfitting measures of
the three optimizers and for all the datasets on which
we conducted our experiments. The results show
that there exits evidence of overfitting happening
during the learning process. Of the three compared
optimizers, GA seems to have suffered the least
from overfitting whereas DE has suffered the most.
We have to point out, however, that for each dataset
Table 2: The overfitting values of DE, PSO, and GA.
Dataset DE PSO GA
Beef 0.12 0.10 0.13
CBF 0.21 0.21 0.19
CinC_ECG_torso 0.27 0.24 0.25
Coffee 0.15 0.14 0.14
ECG200 0.06 0.04 0.01
ECGFiveDays 0.05 0.04 0.05
FaceFour 0.32 0.31 0.17
FISH 0.06 0.06 0.06
Gun_Point 0.11 0.11 0.10
Lighting7 0.10 0.10 0.08
MedicalImages 0.12 0.10 0.07
MoteStrain 0.07 0.00 0.02
OliveOil 0.32 0.17 0.11
OSULeaf 0.08 0.09 0.06
SonyAIBORobotSurfaceII 0.08 0.08 0.08
SwedishLeaf 0.02 0.11 0.10
Symbols 031 0.26 0.24
synthetic_control 0.22 0.15 0.09
Trace 0.10 0.06 0.07
TwoLeadECG 0.16 0.14 0.12
yoga 0.05 0.05 0.00
the three optimizers have suffered from overfitting
in a similar way (to a certain degree), so the
explanation of the results presented in Table 1 can
finally be attributed to differences between the
training sets and the testing sets.
We have to mention here that during the
experiments we also recorded execution time, but
we did not report these results for space restrictions.
The execution time was clearly in favor of PSO,
whereas DE and GA ran at similar speeds.
5 DISCUSSION
There are several counter measures to avoid
overfitting. In (Witten and Frank, 2009) the authors
suggest using a simplest-first search and stopping
when a sufficiently complex concept description is
found. In the language of bio-inspired optimization
this is translated as terminating the search algorithm
earlier, or using fewer parameters. As for the latter
idea, the parameters we are using are intrinsic to the
problem and their number cannot be reduced.
Besides, the number of the parameters in our
problem, 3, is already small. As for the former idea,
we did conduct experiments (which we did not
report here) where NrGen was set to 10 and to 20,
but we did not see a significant difference in
ICPRAM2015-InternationalConferenceonPatternRecognitionApplicationsandMethods
352

performance between the results we obtained for
these values and those we reported in this paper for
which NrGen was set to 100. The reasons for the
overfitting of the solutions resulting from the FCM
clustering might be the nature of the application
itself (Muhammad Fuad, 2013)
6 CONCLUSIONS
The application of bio-inspired optimization
algorithms to data mining tasks is not trivial given
the complexity of these tasks and the stochastic
behavior of bio-inspired algorithms. In this paper we
applied three widely-used bio-inspired optimization
algorithms: differential evolution, genetic
algorithms, and particle swarm optimization, to the
task of fuzzy c-means clustering of time series data,
where the aforementioned optimizers were used to
obtain the optimal values of the weights assigned to
a combination of distance metrics that was used in
the FCM clustering of the time series. We showed in
the experiments we conducted how, while all the
optimizers managed to improve the performance on
the training datasets, the improvement dropped when
the optimized values of the weights were applied to
the testing datasets as a result of the overfitting
problem which appeared during the optimization
process.
REFERENCES
Bustos, B. and Skopal, T., 2006. Dynamic similarity
search in multi-metric spaces. Proceedings of the ACM
Multimedia, MIR Workshop. ACM Press, New York,
NY.
Esling, P., and Agon, C., 2012. Time-series data mining
.ACM Comput. Surv.
Feoktistov, V. , 2006. Differential evolution: in search of
solutions (Springer Optimization and Its
Applications). Secaucus, NJ, USA. Springer- Verlag
New York, Inc.
Krzysztof, J.C., Pedrycz, W., Swiniarski, R.W., Kurgan,
L.A., 2007. Data mining: a knowledge discovery
approach. Springer-Verlag New York, Inc. Secaucus,
New Jersey.
Haupt, R.L., Haupt, S. E., 2004. Practical genetic
algorithms with CD-ROM. Wiley-Interscience.
Keogh, E., Zhu, Q., Hu, B., Hao. Y., Xi, X., Wei, L. &
Ratanamahatana C. A.. 2011.: The UCR time series
classification/clustering homepage: www.cs.ucr.edu/
~eamonn/time_series_data/
Larose, D., 2005. Discovering knowledge in data: an
introduction to data mining, Wiley, Hoboken, NJ.
Liao, T., 2005. Clustering of time series data–a survey.
Pattern Recognition.
Maulik, U., Bandyopadhyay, S., Mukhopadhyay, A.,2011.
Multiobjective genetic algorithms for clustering:
applications in data mining and bioinformatics.
Springer.
Mitchell, M., 1996. An introduction to genetic algorithms,
MIT Press, Cambridge, MA.
Muhammad Fuad, M.M., 2014a. A synergy of artificial
bee colony and genetic algorithms to determine the
parameters of the Σ-Gram distance. In DEXA 2014,
Munich, Germany. Lecture Notes in Computer Science
Volume 8645, 2014, pp 147-154.
Muhammad Fuad, M.M. , 2012a. Differential evolution
versus genetic algorithms: towards symbolic aggregate
approximation of non-normalized time series. In
IDEAS’12 , Prague, Czech Republic, Published by
BytePress/ACM.
Muhammad Fuad, M.M., 2014b. One-step or two-step
optimization and the overfitting phenomenon: a case
study on time series classification. In ICAART 2014.
Muhammad Fuad, M.M., 2012b. Using differential
evolution to set weights to segments with different
information content in the piecewise aggregate
approximation. In KES 2012, San Sebastian, Spain,
(FAIA). IOS Press.
Muhammad Fuad, M.Mm 2013. When optimization is just
an illusion. In ADMA 2013, Part I. LNCS, vol. 8346,
pp. 121–132. Springer, Heidelberg(2013).
Witten, I.H., Frank, E., 2009. Data mining practical
machine learning tools and techniques, second
edition, Elsevier.
OntheApplicationofBio-InspiredOptimizationAlgorithmstoFuzzyC-MeansClusteringofTimeSeries
353
