
Text Mining Technologies for Database Curation
Fabio Rinaldi
Institute of Computational Linguistics, University of Zurich, Zurich, Switzerland
Keywords:
Biomedical Text Mining, Information Extraction, Literature-based Discovery.
Abstract:
Text mining technologies, coupled with advanced user interfaces, have a great potential in the life sciences,
for example supporting the process of database curation. We present a system which has achieved competitive
results in several community-organized evaluations of text mining technologies and we discuss how such
technologies can be integrated in a curation workflow.
1 INTRODUCTION
Biomedical text mining is a discipline that has been
developing quite extensively in recent years. Its aim
is to automatically analyze biomedical text, and in
particular the scientific literature. Several techniques
from the fields of Computational Linguistics, Natu-
ral Language Processing (NLP) and Information Re-
trieval have been adopted for this purpose. Some
of the software tools developed by researchers in
biomedical text mining have the potential to support
the process of database curation, but can of course be
used also for other purposes, such as to enhance the
access to the information contained in the biomedical
literature for different target groups, not only biologi-
cal researchers, but also the general public. Biomedi-
cal text mining is also of great relevance for the phar-
maceutical industry. On average, it costs about 1
billion dollars to develop a completely new medici-
nal drug, and it involves the work of hundreds of re-
searchers, collectively investing in average more than
7 million hours of work on thousands of experiments.
Automated detection of previous information from
the literature can help better target such experiments,
for example by pointing to similar experiments con-
ducted in the past, or suggesting novel experiments on
the basis of previous results. This in turn is going to
have a major economical benefit, freeing up resources
for novel research.
Text mining technologies are increasingly pro-
viding an effective response to the growing demand
for faster access to the vast amounts of informa-
tion hidden in the literature. Several tools are be-
coming available which offer the capability to mine
the literature for specific information, such as for
example protein-protein interactions or drug-disease
relationships. Examples of well known biomedi-
cal text mining tools are MetaMap (Aronson and
Lang, 2010), MedEvi (Kim et al., 2008), WhatIzIt
(Rebholz-Schuhmann et al., 2008), Gimli (Campos
et al., 2013), iHOP (Hoffmann and Valencia, 2004;
Hoffmann, 2007), cTAKES (Savova et al., 2010),
Open Biomedical Annotator (Jonquet et al., 2009).
The biomedical text mining community regularly
verifies the progress of the field through competi-
tive evaluations, such as BioCreative (Arighi et al.,
2011; Krallinger et al., 2011), BioNLP (Kim et al.,
2011; Cohen et al., 2009), i2b2 (Sun et al., 2013),
CALBC (Rebholz-Schuhmann et al., 2011), CLEF-
ER (Rebholz-Schuhmann et al., 2013), DDI (Segura-
Bedmar et al., 2011), BioASQ (Androutsopoulos,
2013), etc. Each of these competitions targets dif-
ferent aspects of the problem, such as detection of
mentions of specific entities (e.g. genes and chem-
icals), detection of protein interactions, assignment
of Gene Ontology tags (BioCreative), detection of
structured events (BioNLP), information extraction
from clinical text (i2b2), large-scale entity detection
(CALBC), multilingual entity detection (CLEF-ER),
drug-drug interactions (DDI), question answering in
biology (BioASQ).
The OntoGene group
1
at the Institute of Computa-
tional Linguistics of the University of Zurich has de-
veloped a platform for advanced text mining applica-
tions. The OntoGene system specializes in the detec-
tion of entities and relationships from selected cate-
gories, such as proteins, genes, drugs, diseases, chem-
icals. OntoGene sources its lexical resources from life
1
www.ontogene.org
544
Rinaldi F..
Text Mining Technologies for Database Curation.
DOI: 10.5220/0005174905440548
In Proceedings of the International Conference on Knowledge Discovery and Information Retrieval (SSTM-2014), pages 544-548
ISBN: 978-989-758-048-2
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

sciences databases, thus allowing a deeper connection
between the unstructured information contained in the
literature and the structured information contained in
databases. The quality of the system has been tested
several times through participation in some of the
community-organized evaluation campaigns, where it
often obtained top-ranked results.
Within the scope of the SNF-funded SASEBio
project (Semi-Automated Semantic Enrichment of the
Biomedical Literature, SNF grant 105315 130558/1,
2010-2014) we developed tools aimed at supporting
the process of database curation from the biomedical
literature, and promote a move towards assisted cu-
ration. By assisted curation we mean a combination
of text mining approaches and the work of an expert
curator, aimed at leveraging the power of text mining
systems, while retaining the high quality associated
with human expertise.
In the rest of this paper, we briefly describe in
Section 2 the overall architecture of our text mining
system and mention evaluation results in the context
of community-organized shared tasks, then we illus-
trate in Section 3 the usage of the text mining system
within our integrated curation environment, providing
a discussion on assisted curation and results of collab-
orations with major life science databases.
2 THE OntoGene TEXT MINING
SYSTEM
In this section we provide a brief description of the
OntoGene text mining environment, with a specific
focus on its application to the detection of interac-
tions. The first step in order to process a collec-
tion of biomedical literature consists in the annota-
tion of names of relevant domain entities in biomed-
ical literature (we consider in particular proteins,
genes, species, experimental methods, and cell lines)
and grounding them to widely accepted identifiers
(IDs) assigned by reference knowledge bases, such as
UniProt, EntrezGene, Cell Line Knowledge Base, etc.
The term annotation uses a large term list that is com-
piled on the basis of the entity names extracted from
the knowledge bases. This resulting list covers the
common expression of the terms. A term normaliza-
tion step is used to match the terms with their actual
representation in the text, taking into account a num-
ber of possible surface variations. Finally, a disam-
biguation step resolves the ambiguity (i.e. multiple
IDs proposed by the annotator) of the matched terms
(Rinaldi et al., 2011).
In order to account for possible surface variants
between the terms in the term list and the token se-
quences in the text, a normalization step is included in
the annotation procedure. The same normalization is
applied to the known terms of the term list and to the
candidate terms in the input text, so that a matching
between variants becomes possible despite the differ-
ences in the surface strings. In case the normalized
strings match exactly, the input sequence is annotated
with the IDs of the matching term.
Using the information concerning mentions of rel-
evant domain entities, derived as described above,
and their corresponding unique identifiers obtained by
the process of disambiguation, it is possible to cre-
ate candidate interactions using the co-occurrence of
two entities in a given text span (typically a sentence,
or observation window). However, using simple co-
occurrence leads to low-precision extraction of inter-
actions. In order to obtain better precision it is neces-
sary to take into account the syntactic structure of the
sentence, and other structural information. We parse
relevant sentences (containing at least two entities)
with a dependency parser, which has been adapted
to and evaluated on the biomedical domain (Schnei-
der et al., 2007). After parsing, we collect all syn-
tactic connections that exist between all the terms as
follows. For each term-coocurrence a collector tra-
verses the tree from one term up to the lowest com-
mon mother node, and down the second term, record-
ing all intervening nodes. Paths which are extracted
from the corpus can directly be used for interaction
detection. The ranking of relation candidates can be
further optimized if we apply a supervised machine
learning method. First we automatically identify the
noisy concepts that our term recognizer generates in
order to penalize them. Second, we need to adapt to
highly-ranked false positive relations which are gen-
erated by our frequency based approach. The goal is
to identify some global preference oder biases which
can be found in the reference database.
We have been active in the area of biomedical text
mining since 2005, participating in several compet-
itive evaluations, and often obtaining top-ranked re-
sults. Some significant examples are the following:
best results in finding mentions of experimental meth-
ods in BioCreative 2006 (Rinaldi et al., 2008), best re-
sults in detecting protein-protein interactions from the
literature in BioCreative 2009 (Rinaldi et al., 2010),
best results in detecting some entity types (genes and
diseases in particular) in the CALBC competition
(Rebholz-Schuhmann et al., 2010), best overall re-
sults in the triage task of BioCreative 2012 (Rinaldi
et al., 2013a). In addition we have been actively pro-
moting the idea that advanced text mining technolo-
gies can be a helpful support tool within the context of
a curation workflow, as described in the next section.
TextMiningTechnologiesforDatabaseCuration
545
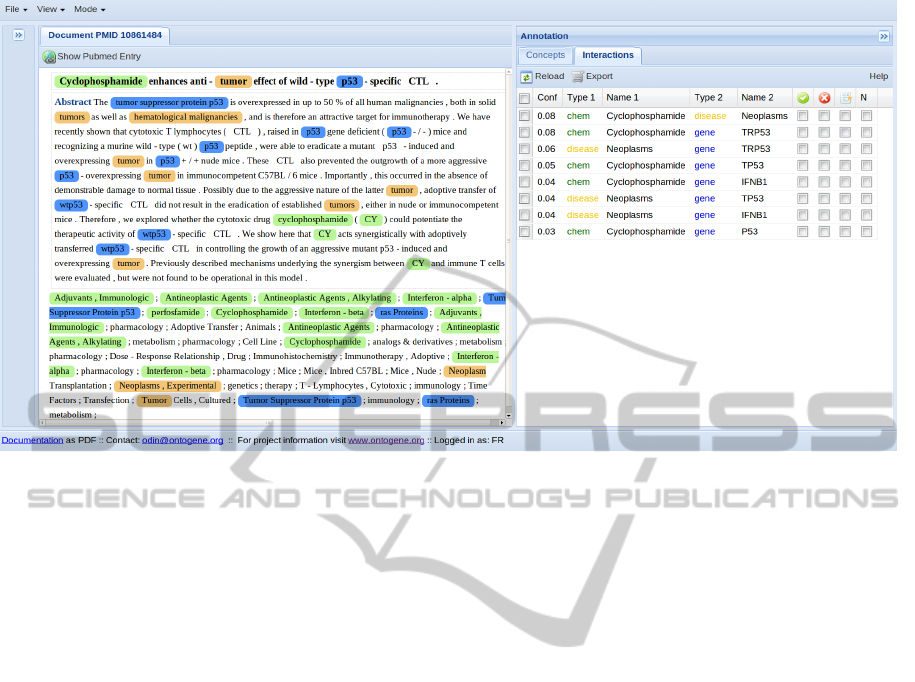
Figure 1: ODIN screenshot. Example of visualization of text mining results using the ODIN interface. The panel on the left
shows the document with annotations, the panel on the right the corresponding concepts. The two panels are interconnected
by the interface logic: whenever an item is selected in the concept panel, the corresponding terms are highlighted in the
document panel.
3 ASSISTED CURATION
The main motivation behind biomedical curation ac-
tivities is “to help the life sciences community make
sense of all the data that is accumulating” (Bairoch,
2009). Although human curation offers the best guar-
antee of high quality results, it suffers from severe
bottlenecks which have long been recognized in the
curation community. The most pressing problem is
that of efficiency of the process: despite the fact that
typically several databases attempt to focus on a par-
ticular type of biological data, and often collaborate at
least sufficiently to prevent duplication of effort and
ensure compatibility of resulting data formats, it is
impossible for human curators to keep up with the
growing pace of publication: “Nobody will ever be
able to manually annotate all the macromolecular bi-
ological entities that exist on this planet, and conse-
quently automatization is the only solution” (Bairoch,
2009).
However, automated text mining tools cannot of-
fer sufficient reliability to be applied indiscriminately
without human supervision. Therefore, the ideal solu-
tion is to combine the best capabilities of automated
systems with human supervision by highly qualified
domain experts. For this type of application, it is
necessary to develop user friendly interfaces that will
make text mining tools directly usable by curators,
rather than hinder their work with technical com-
plexities and poorly presented results. The Onto-
Gene group has implemented a user-friendly curation
framework called ODIN (Ontogene Document IN-
spector, see figure 1), which aims at satisfying several
of these requirements. Since every curation group has
specific interests and needs, it cannot be expected that
generic text mining solutions will be able to provide a
satisfactory solution to all of them. Instead, we intend
to provide a generic framework that can be then cus-
tomized to the specific requirements of each group.
The usage of ODIN as a curation tool has been
tested in a few collaborations with curation groups,
including PharmGKB (Rinaldi et al., 2012a), CTD
(Rinaldi et al., 2012b), RegulonDB (Gama-Castro
et al., 2013; Rinaldi et al., 2013b; Gama-Castro et al.,
2014), which are briefly described here.
PharmGKB (The Pharmacogenomics Knowledge
Base) (Sangkuhl et al., 2008) is a NIH-funded, pub-
licly available online resource, developed and main-
tained at Stanford University, which aims at curating
information pertaining to the effect of genetic vari-
ants in susceptibility to diseases and drugs. They cu-
rate publications which are related to genetic variants,
and they annotate interactions between genes, drugs
and diseases. In 2011 we performed an experiment
in assisted curation in collaboration with PharmGKB,
which demonstrated the high usability level of ODIN
KDIR2014-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
546

for a real-world curation task, as well as proved the
quality of the results of the underlying text mining al-
gorithms (Rinaldi et al., 2012a).
In 2012, as part of our participation in the BioCre-
ative 2012 text mining evaluation campaign, we an-
alyzed data provided by the Comparative Toxicoge-
nomics Database (CTD) (Davis et al., 2011), which
aims at collecting information related to the health ef-
fects of environmental chemicals. They curate rela-
tionships among chemicals, genes and diseases. We
adapted our text mining pipeline to their specific pur-
poses, and created for them a customized version of
our ODIN tool. Once again, we obtained the best re-
sults in the competition, as described in detail in (Ri-
naldi et al., 2013a).
RegulonDB (Gama-Castro et al., 2011) is another
major biological database, focusing on regulatory in-
teractions of one model organism (E. coli). Regu-
lonDB is the primary reference database of the best-
known regulatory network of any free-living organ-
ism. A collaboration between OntoGene and Regu-
lonDB was initiated in 2013 and resulted in a joint
participation in the BioCreative 2013 interactive cura-
tion task. Once again, ODIN was customized for the
specific needs of the RegulonDB database and was
tested by RegulonDB curators. Novel sentence fil-
ters were implemented, which allow the curators to
see only the sentences which satisfy a given logical
condition. The joint experiment described in (Gama-
Castro et al., 2014) showed that the usage of such fil-
ters allows an expert curator to reduce the amount of
text material under consideration by a factor of 10,
without any loss of accuracy in the curated results.
4 CONCLUSION
We have presented an advanced text mining architec-
ture (OntoGene Text Miner), which is embedded in
a user-friendly curation interface (ODIN). The Onto-
Gene Text Miner has been evaluated in a number of
competitive evaluation tasks and shown to perform at
state-of-the-art levels.
Besides, it has already been tested in a number of
real-world curation tasks in collaboration with major
life sciences databases.
ACKNOWLEDGEMENTS
The OntoGene group is partially supported by the
Swiss National Science Foundation (grants 100014 −
118396/1 and 105315 − 130558/1) and by Hoffman-
La Roche Pharmaceuticals, Basel, Switzerland.
REFERENCES
Androutsopoulos, I. (2013). A challenge on large-scale
biomedical semantic indexing and question answer-
ing. In BioNLP workshop (part of the ACL Confer-
ence).
Arighi, C., Roberts, P., Agarwal, S., Bhattacharya, S.,
Cesareni, G., Chatr-aryamontri, A., Clematide, S.,
Gaudet, P., Giglio, M., Harrow, I., Huala, E.,
Krallinger, M., Leser, U., Li, D., Liu, F., Lu, Z., Mal-
tais, L., Okazaki, N., Perfetto, L., Rinaldi, F., Sae-
tre, R., Salgado, D., Srinivasan, P., Thomas, P., Toldo,
L., Hirschman, L., and Wu, C. (2011). Biocreative iii
interactive task: an overview. BMC Bioinformatics,
12(Suppl 8):S4.
Aronson, A. R. and Lang, F. M. (2010). An overview of
MetaMap: historical perspective and recent advances.
J Am Med Inform Assoc, 17(3):229–236.
Bairoch, A. (2009). The future of annotation/biocuration.
Nature Precedings.
Campos, D., Matos, S., and Oliveira, J. L. (2013). Gimli:
open source and high-performance biomedical name
recognition. BMC Bioinformatics, 14:54.
Cohen, K. B., Demner-Fushman, D., Ananiadou, S., Pes-
tian, J., Tsujii, J., and Webber, B., editors (2009). Pro-
ceedings of the BioNLP 2009 Workshop. Association
for Computational Linguistics, Boulder, Colorado.
Davis, A., King, B., Mockus, S., Murphy, C., Saraceni-
Richards, C., Rosenstein, M., Wiegers, T., and
Mattingly, C. (2011). The comparative toxicoge-
nomics database: update 2011. Nucleic Acids Res.,
39(Database issue):D1067–72.
Gama-Castro, S., Rinaldi, F., Lpez-Fuentes, A., Balderas-
Martnez, Y. I., Clematide, S., Ellendorff, T. R., and
Collado-Vides, J. (2013). Assisted curation of growth
conditions that affect gene expression in e. coli k-12.
In Proceedings of the Fourth BioCreative Challenge
Evaluation Workshop, volume 1, pages 214–218.
Gama-Castro, S., Rinaldi, F., Lpez-Fuentes, A., Balderas-
Martnez, Y. I., Clematide, S., Ellendorff, T. R.,
Santos-Zavaleta, A., Marques-Madeira, H., and
Collado-Vides, J. (2014). Assisted curation of regu-
latory interactions and growth conditions of OxyR in
E. coli K-12. Database: The Journal of Biological
Databases and Curation, bau049.
Gama-Castro, S., Salgado, H., Peralta-Gil, M., Santos-
Zavaleta, A., Muniz-Rascado, L., Solano-Lira, H.,
Jimenez-Jacinto, V., Weiss, V., Garcia-Sotelo, J. S.,
Lopez-Fuentes, A., Porron-Sotelo, L., Alquicira-
Hernandez, S., Medina-Rivera, A., Martinez-Flores,
I., Alquicira-Hernandez, K., Martinez-Adame, R.,
Bonavides-Martinez, C., Miranda-Rios, J., Huerta,
A. M., Mendoza-Vargas, A., Collado-Torres, L.,
Taboada, B., Vega-Alvarado, L., Olvera, M., Olvera,
L., Grande, R., Morett, E., and Collado-Vides, J.
(2011). RegulonDB version 7.0: transcriptional reg-
ulation of Escherichia coli K-12 integrated within ge-
netic sensory response units (Gensor Units). Nucleic
Acids Res., 39(Database issue):98–105.
Hoffmann, R. (2007). Using the iHOP information resource
to mine the biomedical literature on genes, proteins,
TextMiningTechnologiesforDatabaseCuration
547

and chemical compounds. Curr Protoc Bioinformat-
ics, Chapter 1:Unit1.16.
Hoffmann, R. and Valencia, A. (2004). A gene network for
navigating the literature. Nature Genetics, 36:664.
Jonquet, C., Shah, N. H., and Musen, M. A. (2009). The
open biomedical annotator. Summit on Translat Bioin-
forma, 2009:56–60.
Kim, J., Pezik, P., and Rebholz-Schuhmann, D. (2008).
Medevi: Retrieving textual evidence of relations be-
tween biomedical concepts from medline. Bioinfor-
matics, 24(11):1410–1412.
Kim, J., Pyysalo, S., Ohta, T., Bossy, R., Nguyen, N., and
Tsujii, J. (2011). Overview of bionlp shared task 2011.
ACL HLT 2011, page 1.
Krallinger, M., Vazquez, M., Leitner, F., Salgado, D., Chatr-
aryamontri, A., Winter, A., Perfetto, L., Briganti, L.,
Licata, L., Iannuccelli, M., Castagnoli, L., Cesareni,
G., Tyers, M., Schneider, G., Rinaldi, F., Leaman, R.,
Gonzalez, G., Matos, S., Kim, S., Wilbur, W., Rocha,
L., Shatkay, H., Tendulkar, A., Agarwal, S., Liu, F.,
Wang, X., Rak, R., Noto, K., Elkan, C., Lu, Z., Dogan,
R., Fontaine, J.-F., Andrade-Navarro, M., and Valen-
cia, A. (2011). The protein-protein interaction tasks
of biocreative iii: classification/ranking of articles and
linking bio-ontology concepts to full text. BMC Bioin-
formatics, 12(Suppl 8):S3.
Rebholz-Schuhmann, D., Arregui, M., Gaudan, S., Kirsch,
H., and Jimeno, A. (2008). Text processing through
Web services: calling Whatizit. Bioinformatics,
24(2):296–298.
Rebholz-Schuhmann, D., Clematide, S., Rinaldi, F.,
Kafkas, S., van Mulligen, E. M., Bui, C., Hellrich,
J., Lewin, I., Milward, D., Poprat, M., Jimeno-Yepes,
A., Hahn, U., and Kors, J. (2013). Entity recogni-
tion in parallel multi-lingual biomedical corpora: The
clef-er laboratory overview. In Forner, P., Mueller, H.,
Rosso, P., and Paredes, R., editors, Information Access
Evaluation. Multilinguality, Multimodality, and Visu-
alization, Lecture Notes in Computer Science, pages
353–367. Springer, Valencia.
Rebholz-Schuhmann, D., Jimeno, A., Li, C., Kafkas,
S., Lewin, I., Kang, N., Corbett, P., Milward, D.,
Buyko, E., Beisswanger, E., Hornbostel, K., Alex,
Kouznetsov, r., Witte, R., Laurila, J. B., Baker, C. J.,
Chen-Kuo, J., Clematide, S., Rinaldi, F., Farkas, R.,
M
´
ora, G., Hara, K., Furlong, L., Rautschka, M.,
Neves, M. L., Pascual-Montano, A., Wei, Q., Col-
lier, N., Chowdhury, F. M., Lavelli, A., Berlanga, R.,
Morante, R., Van Asch, V., Daelemans, W., Marina,
J. L., Mulligen, E. v., Kors, J., and Hahn, U. (2010).
Assessment of NER solutions against the first and sec-
ond CALBC Silver Standard Corpus. In Semantic
Mining in Medicine, EBI, Cambridge, UK,.
Rebholz-Schuhmann, D., Yepes, A., Li, C., Kafkas, S.,
Lewin, I., Kang, N., Corbett, P., Milward, D., Buyko,
E., Beisswanger, E., Hornbostel, K., Kouznetsov, A.,
Witte, R., Laurila, J., Baker, C., Kuo, C.-J., Clematide,
S., Rinaldi, F., Farkas, R., Mora, G., Hara, K.,
Furlong, L. I., Rautschka, M., Neves, M., Pascual-
Montano, A., Wei, Q., Collier, N., Chowdhury, M.,
Lavelli, A., Berlanga, R., Morante, R., Van Asch, V.,
Daelemans, W., Marina, J., van Mulligen, E., Kors,
J., and Hahn, U. (2011). Assessment of ner solu-
tions against the first and second calbc silver standard
corpus. Journal of Biomedical Semantics, 2(Suppl
5):S11.
Rinaldi, F., Clematide, S., Garten, Y., Whirl-Carrillo, M.,
Gong, L., Hebert, J. M., Sangkuhl, K., Thorn, C. F.,
Klein, T. E., and Altman, R. B. (2012a). Using ODIN
for a PharmGKB re-validation experiment. Database:
The Journal of Biological Databases and Curation.
Rinaldi, F., Clematide, S., and Hafner, S. (2012b). Rank-
ing of ctd articles and interactions using the onto-
gene pipeline. In Proceedings of the 2012 BioCreative
workshop, Washington D.C.
Rinaldi, F., Clematide, S., Hafner, S., Schneider, G.,
Grigonyte, G., Romacker, M., and Vachon, T. (2013a).
Using the OntoGene pipeline for the triage task
of BioCreative 2012. The Journal of Biological
Databases and Curation, Oxford Journals.
Rinaldi, F., Gama-Castro, S., Lpez-Fuentes, A., Balderas-
Martnez, Y., and Collado-Vides, J. (2013b). Digital
curation experiments for regulondb. In BioCuration
2013, April 10th, Cambridge, UK.
Rinaldi, F., Kaljurand, K., and Saetre, R. (2011). Termi-
nological resources for text mining over biomedical
scientific literature. Journal of Artificial Intelligence
in Medicine, 52(2):107–114.
Rinaldi, F., Kappeler, T., Kaljurand, K., Schneider, G.,
Klenner, M., Clematide, S., Hess, M., von Allmen,
J.-M., Parisot, P., Romacker, M., and Vachon, T.
(2008). OntoGene in BioCreative II. Genome Biol-
ogy, 9(Suppl 2):S13.
Rinaldi, F., Schneider, G., Kaljurand, K., Clematide, S., Va-
chon, T., and Romacker, M. (2010). OntoGene in
BioCreative II.5. IEEE/ACM Transactions on Com-
putational Biology and Bioinformatics, 7(3):472–480.
Sangkuhl, K., Berlin, D. S., Altman, R. B., and Klein, T. E.
(2008). PharmGKB: Understanding the effects of in-
dividual genetic variants. Drug Metabolism Reviews,
40(4):539–551. PMID: 18949600.
Savova, G. K., Masanz, J. J., Ogren, P. V., Zheng, J., Sohn,
S., Kipper-Schuler, K. C., and Chute, C. G. (2010).
Mayo clinical Text Analysis and Knowledge Extrac-
tion System (cTAKES): architecture, component eval-
uation and applications. J Am Med Inform Assoc,
17(5):507–513.
Schneider, G., Kaljurand, K., Rinaldi, F., and Kuhn, T.
(2007). Pro3Gres parser in the CoNLL domain adap-
tation shared task. In Proceedings of the CoNLL
Shared Task Session of EMNLP-CoNLL 2007, pages
1161–1165, Prague.
Segura-Bedmar, I., Martnez, P., and Snchez-Cisneros, D.
(2011). The 1st ddi extraction-2011 challenge task:
Extraction of drug-drug interactions from biomedical
texts. In Proc DDI Extraction-2011 challenge task,
pages 1–9, Huelva, Spain.
Sun, W., Rumshisky, A., and Uzuner, O. (2013). Evaluating
temporal relations in clinical text: 2012 i2b2 Chal-
lenge. J Am Med Inform Assoc, 20(5):806–813.
KDIR2014-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
548
