
Generic 3D Segmentation in Medicine based on a Self-learning
Topological Model
Gerald Zwettler
1
and Werner Backfrieder
1,2
1
Bio - and Medical Informatics, Research and Development Department, Upper Austria University of Applied Sciences,
Softwarepark 11, 4232 Hagenberg, Austria
2
School of Informatics, Communication and Media, Upper Austria University of Applied Sciences,
Softwarepark 11, 4232 Hagenberg, Austria
Keywords: Model-based Image Segmentation, Statistical Image Classification, Hybrid Watershed Pre-segmentation.
Abstract: Three-dimensional segmentation of medical image data is crucial in modern diagnostics and still subject of
intensive research efforts. Most fully automated methods, e.g. the segmentation of the hippocampus, are
highly specific for certain morphological regions and very sensitive to variations in input data, thus
robustness is not sufficient to achieve sufficient accuracy to serve in differential diagnosis. In this work a
processing pipeline for robust segmentation is presented. The flexibility of this novel generic segmentation
method is based on entirely parameter-free pre-segmentation. Therefore a hybrid modification of the
watershed algorithm is developed, employing both gradient and intensity metrics for the identification of
connected regions depending on similar properties. In a further optimization step the vast number of small
regions is condensed to anatomically meaningful structures by feature based classification. The core of the
classification process is a topographical model of the segmented body region, representing a sufficient
number of features from geometry and the texture domain. The model may learn from manual segmentation
by experts or from its own results. The novel method is demonstrated for the human brain, based on the
reference data set from brainweb. Results show high accuracy and the method proves to be robust. The
method is easily extensible to other body regions and the novel concept shows high potential to introduce
generic segmentation in the three-dimensional domain into a clinical work-flow.
1 INTRODUCTION
The accurate and preferably fully-automated
segmentation of medical image data is of high
importance for a broad range of medical
applications. The importance of computer-based
support for surgery planning, disease monitoring and
general diagnostics, by allowing for precise
estimation of volume, size and relative position of
anatomical structures, will constantly grow in
clinical practice. As an example, after automated
segmentation of liver parenchyma, hepatic vessels
and possible lesions utilizing level sets, the tumour
position can be analyzed with respect to the
supporting vessels and liver lobes which is of high
importance for surgery planning (Zwettler et al.,
2009). The informative value image data from the
functional imaging domain like SPECT or PET can
be raised by combining high anatomical resolution
of tomographic modalities like MRI and CT.
Thereby, the metabolic activity can be quantified
utilizing patient specific segmentation masks derived
from the anatomical imaging (Beyer et al., 2010).
In this work we present a concept for model-
based segmentation of 3D tomographic medical
image data based on a generic, parameter-free pre-
segmentation process and texture feature driven
region merging for classification. Our novel pre-
segmentation strategy combines aspects of gradient
based watershed transform (Beare and Lehmann,
2006), confidence connected region growing, region
merging and new variations in a hybrid algorithm
(Zwettler and Backfrieder, 2012). Starting at local
minima positions besides gradient height of classical
watershed transform, region intensity statistics are
used as merge metric. For region merging of the
initially pre-processed regions, several metrics,
namely watershed level tolerance, geometric
properties and similarity of the intensity profile are
combinded. Thus, an arbitrary input image can be
104
Zwettler G. and Backfrieder W..
Generic 3D Segmentation in Medicine based on a Self-learning Topological Model.
DOI: 10.5220/0004294701040108
In Proceedings of the International Conference on Computer Vision Theory and Applications (VISAPP-2013), pages 104-108
ISBN: 978-989-8565-48-8
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)
pre-classified at a user specified number of target
regions, defining the granularity of this pre-
processing step. Based on the pre-segmented
regions, the final segmentation is achieved by
feature-based classification utilizing cost
optimization. Besides texture features, also
geometric properties are incorporated, all derived
from a statistical a priori model calculated from
manual reference segmentations. For precise
reference segmentations at low user interactions,
rapid prototyping image processing chains have
been evaluated. The graph-based topographic
modelling of the anatomical context to segment
allows the segmentations at different hierarchical
levels. Applicability for future multi-modal image
processing will be evaluated in future.
2 DATA
For testing purposes concerning manual reference
segmentations, automated pre-segmentation and
feature-based classification, n=20 T1-weighted MRI
datasets from the simulated brainweb database
(Kwan et al., 1999) and the associated reference
segmentations are used.
Further test runs and validations are performed
utilizing n=12 anonymous multi-modal patient
studies, comprising morphologic image acquisitions
(T1, T2, PD,...) as well as related functional imaging
(SPECT, PET). For the patient data sets, the required
reference segmentations required for model training
and leave-one-out validation are achieved in a semi-
automated way by applying image processing
pipelines utilizing MeVisLab modules (Ritter, 2007)
a discussed in the later sections.
3 METHODOLOGY
In the preparation phase of our segmentation
concept, model definition is performed with respect
to the imaging modality to support and the
hierarchical anatomical topography of interest.
Furthermore, the classification features are chosen
with respect to their correlation. Based on the
anatomical topography and the chosen features, a
sufficient set of reference segmentations has to be
processed applying a semi-automated image
processing chain to evaluate the model parameters
with respect to each particular feature and each
particular anatomical structure to consider.
After preparing and training the model,
pre-segmentation of the tomographic patient dataset
to process is performed, utilizing a hybrid approach
incorporating aspects of watershed transform,
confidence connected region growing and region
merging. The chosen features are evaluated for all
regions resulting from the pre-segmentation process
step. The final segmentation is interpreted as
optimization problem, as the anatomical structure
labels are assigned to the particular regions in a way
to achieve a classification result that minimizes the
overall error with respect to the statistical feature
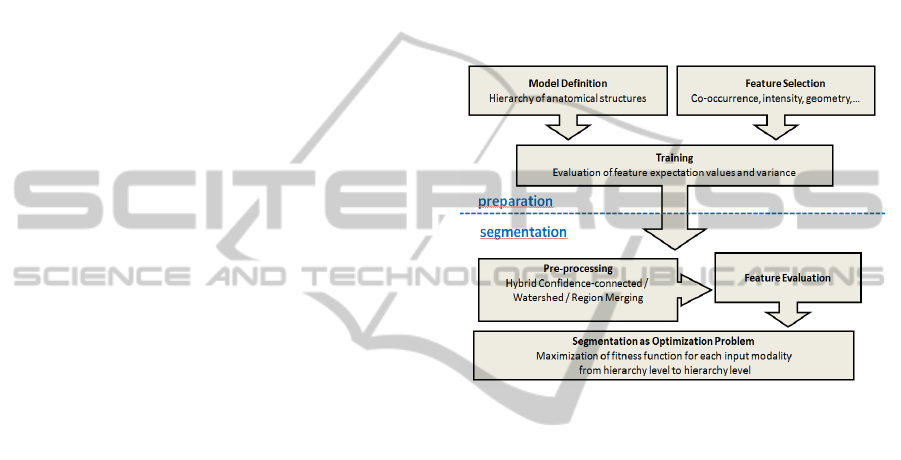
values, see Fig 1.
Figure 1: Illustration of our model-based segmentation
strategy. After definition of anatomical topography and
model training as preparation, pre-segmentation of the 3D
tomographic data is performed and resulting regions are
classified according to statistical features for final
segmentation.
3.1 Definition of Hierarchical
Anatomical Topography
For each anatomical structure to segment in the
particular medical context and for the specified
imaging modality, the position within the cascading
hierarchy of granularity has to be defined, see Fig. 2.
At level 0 the first separation into foreground as the
region of interest and the background as remaining
voxels is performed. Later the structures are
subdivided into composing structures according to
anatomical topography.
Generic3DSegmentationinMedicinebasedonaSelf-learningTopologicalModel
105
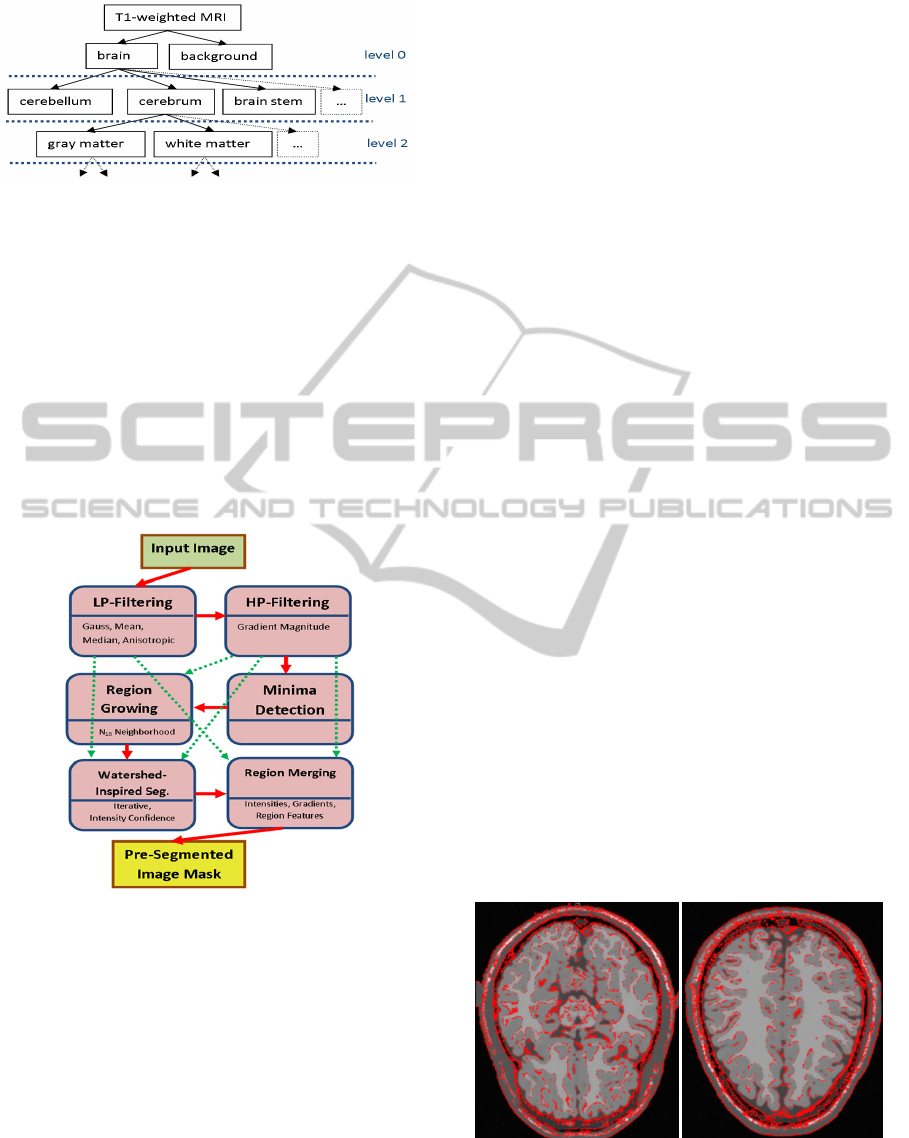
Figure 2: Illustration of topographic modelling.
Granularity is increased at higher levels.
3.2 Hybrid Pre-Segmentation
A generic segmentation of arbitrary image data can
be achieved utilizing an image processing chain,
including anisotropic diffusion filtering for
smoothing image data, gradient magnitude
extraction and a hybrid watershed implementation,
incorporating intensity homogeneity besides
gradient borders and neighbour region boundary
conditions as explained in (Zwettler and
Backfrieder, 2012) and illustrated in Fig. 3.
Figure 3: The sequential process chain (solid red arrows)
comprises low-pass filtering, high-pass filtering, minima
detection region growing, the watershed-type
segmentation procedure and finally region merging until
the convergence criterion is reached. Filtered input image
and the gradient magnitude representation are required as
input for several particular process steps (dashed green
arrows).
3.3 Feature-based Classification
Our segmentation strategy is defined as hierarchical
multi-feature optimization problem for classification
of pre-defined anatomical structures with respect to
their statistical feature values. For each anatomical
object at each hierarchy level, a statistical feature
vector must be preserved as a-priori information.
The chosen features comprise classic metrics
derived from co-occurrence matrix, like entropy and
energy (Felipe, 2003), the deviation of the intensity
profile, results of structural analysis with Hessian
eigenvalues (Sato et al., 1998) and others. Beside
these textural parameters, shape information, like
deviation of the size and extent is incorporated.
The segmentation itself is now defined as an
optimization problem of assigning the pre-classified
3D elements to the defined objects at lowest
hierarchy level to minimize the cumulated total
feature error. Due to recursive dependencies, only
one hierarchy level is optimized at each time and the
results are used as input for the next level in an
iterative top-down and bottom-up cycle. As first
initialization, the best matching regions for the
particular anatomical structures are iteratively
assigned with respect to expected size of the
particular structure. Results after this first
classification run are currently our final results.
In future, refinement of the segmentation results
is expected to be achieved utilizing heuristic
optimization based on evolution strategy
(Rechenberg, 1973) and classic genetic algorithms
(Goldberg, 1989).
4 RESULTS
Utilizing hybrid pre-segmentation, precision of
common watershed segmentation can be increased
from 0.88 to 0.91 and 0.92 respectively for our static
and adaptive intensity interval region merging on
average. The target number of regions to be pre-
segmented is reliably reached and misclassifications
only occur in border areas between neighbouring
anatomical structures, see Fig. 4.
Figure 4: Misclassified voxel (red) of AII4_WS
segmentation strategy in slices #70 and #150 compared to
brainweb reference segmentations displayed with respect
to original image data of dataset subject 04.
VISAPP2013-InternationalConferenceonComputerVisionTheoryandApplications
106

Concerning classification, for each feature the
prediction reliability is calculated based on
distributions with mean μ and variance δ calculated
as statistical average of all reference segmentations
and each particular anatomical structure to
distinguish. The reliability thereby indirectly
correlates with the error due to overlapping areas.
The overall prediction reliability is calculated via
stepwise integration over the entire feature value
range. The distributions of the anatomical structures
are normalized with respect to their overall
probability, i.e. statistical differences in anatomical
structure size.
Currently we evaluate 34 different feature types
for their prediction reliability when training an
anatomical topography model. Some of the most
notable are presented in Table 1 for analysis on
brainweb datasets. For construction of the feature
vector, a number of 10 of the best discriminating
features, should be selected. Despite choosing the
features with highest prediction reliability, it has to
be assured, that correlation within the feature vector
is low.
Table 1: Different types of features and their reliability for
classifying the different anatomical structure feature value
distributions.
feature
ID
description prec.
1 maximum intensity value 67.93
3 median intensity value 87.07
4 mean intensity value 88.43
5 quantile 25 intensity 90.37
7 anatomical structure size 82.34
21 surface-to-volume-ratio 82.29
22 entropy of intensities 75.40
23 energy of intensities 74.12
25 mean probability of intensities 78.46
5 CONCLUSIONS
A generic segmentation concept for fast model-
based adjustment to particular image segmentation
tasks and imaging modalities has been presented.
Hybrid pre-segmentation is perfectly applicable for
context-free pre-processing of arbitrary image data
for first region labelling. Correlation of the analyzed
texture and geometric features shows promising
results for future heuristics-based classification
according to pre-defined anatomical topography.
The discussed and continuously refined rapid
prototyping image processing chain is perfectly
applicable for fast and robust preparation of
reference segmentations for training the a priori
model.
ACKNOWLEDGEMENTS
Thanks to our medical partners from the Wagner-
Jauregg state mental hospital, Linz, Upper Austria,
at the institute for neuro-nuclear medicine headed by
Primarius Dr.Dr. Robert Pichler for providing
medical image data and for valuable discussion.
This research is part of the INVERSIA project
(http://inversia.fh-linz.at) which was funded by the
European Regional Development Fund (ERDF) in
cooperation with the Upper Austria state
government (Regio13).
REFERENCES
Beare, R., and Lehmann, G., 2006. The watershed
transform in ITK – discussion and new developments.
In Insight Journal.
Beyer, T., Schwenzer, N., Bisdas, S., Claussen C.D., and
Pichler, B.J., 2010. MR/PET – Hybrid Imaging for the
Next Decade. In MAGNETOM Flash 3/2010.
Felipe, J. C., 2003. Retrieval by content of medical images
using texture for tissue identification. In CBMS.
Goldberg, D. E., 1989. Genetic Algorithms in Search
Optimization and Machine Learning. In Addison-
Wesley Professional.
Kwan, R. K.-S., Evans, A. C., Pike, G.B., 1999. MRI
simulation-based evaluation of image-processing and
classification methods. In IEEE Transactions on
Medical Imaging 18(11):1085-1097.
Rechenberg, I., 1973. Evolutionsstrategie-Optimierung
technischer Systeme nach Prinzipien der biologischen
Evolution. In Frommann-Holzboog Verlag, Stuttgart,
Germany.
Ritter, F., 2007. Visual Programming for Prototyping of
Medical Applications. In IEEE Visualization
workshop.
Sato, Y. S., Atsumi, H., Koller, T., Gerig, G., Yoshida, S.,
Kikinis, R., 1998. Three-dimensional multi-scale line
filter for segmentation and visualization curvilinear
structures in medical images. In Medical Image
Analysis 2 (2), 143-168.
Zwettler, G., Backfrieder, W., Swoboda, R., Pfeifer, F.,
2009. Fast Fully-automated Model-driven Liver
Segmentation Utilizing Slicewise Applied Levelsets
on Large CT Data. In Proc. of the 21st European
Modeling and Simulation Symposium, 161-166.
Generic3DSegmentationinMedicinebasedonaSelf-learningTopologicalModel
107

Zwettler, G., Backfrieder, W., 2012. A New Hybrid
Algorithm Based on Watershed Method, Confidence
Connected Thresholding and Region Merging as
Preprocessing for Statistical Classification of General
Medical Images. In Proc. of the 24th European
Modeling and Simulation Symposium.
VISAPP2013-InternationalConferenceonComputerVisionTheoryandApplications
108
