
REPRESENTING THE INTERNATIONAL CLASSIFICATION
OF DISEASES VERSION 10 IN OWL
Manuel M
¨
oller, Michael Sintek, Ralf Biedert, Patrick Ernst, Andreas Dengel
German Research Center for Artificial Intelligence (DFKI) and University of Kaiserslautern, Kaiserslautern, Germany
Daniel Sonntag
German Research Center for Artificial Intelligence (DFKI), Saarbrcken, Germany
Keywords:
Formal knowledge representation, Automatic ontology generation, Medical ontologies, International classifi-
cation of diseases.
Abstract:
Current efforts in the biomedical ontology community focus on establishing interoperability and data inte-
gration. In covering human diseases, one of the major international standards in clinical practice is the In-
ternational Classification for Diseases (ICD), maintained by the World Health Organization (WHO). Several
country- and language-specific adaptations exist which share the general structure of the WHO version but
differ in certain details. This complicates the exchange of patient records and hampers data integration across
language borders. We present our approach for modeling the hierarchy of the ICD-10 using the Web Ontology
Language (OWL). Our model captures the hierarchical information of the ICD-10 as well as comprehensive
class labels for English and German. Specialties such as “Exclusion” statements, which make statements about
the disjointness of certain ICD-10 categories, are modeled in a formal way. For properties which exceed the
expressivity of OWL-DL, we provide a separate OWL-Full component which allows us to use the hierarchi-
cal knowledge and class labels with existing OWL-DL reasoners and capture the additional information in a
machine-interpretable way.
1 INTRODUCTION
Over the last decades healthcare has changed from
isolated treatments towards a distributed treatment
process. This process depends greatly on the coop-
eration of specialized medical disciplines. Moreover,
medicine questions require to take into account an
enormous amount of expert knowledge before deci-
sions are made. To facilitate information exchange
and sharing of knowledge in medical domains, stan-
dardization is playing an important role. The goal is
to increase the interoperability within all domains of
the healthcare industry so that the interchange of doc-
uments can be simplified and work flows can be im-
proved.
The difficulty in the area of medical knowledge
management is the high diversity of knowledge about
single entities. Let us consider a patient in a clini-
cal environment. Even if he has a simple disease, he
would have to undergo a high number of examinations
in different clinical departments. In each step of his
treatment, huge amounts of metadata are created and
stored, based on single specific models every time.
The challenge is to integrate these islands of informa-
tion (Lenz, 2005), so that an overall knowledge base
can emerge.
Another problem of this integration process is se-
mantic heterogeneity, which means, that there are dis-
agreements about the semantics or interpretation of
concepts between the terminologies. Additionally,
medical knowledge is very complex and evolves con-
tinuously over time. Therefore, new architectures and
standards are needed that deal with these problems
(Sonntag et al., 2009), (Sonntag, 2010).
Standardized terminologies have a long history in
medicine. For human diseases, the first approaches
date back to the 18th century. The roots of the modern
International Classification of Diseases (ICD) can be
traced back to the Bertillon Classification of Causes
of Death. The ICD was introduced in 1893 at the
International Statistical Institute in Chicago. Five
years later, the American Public Health Association
50
Möller M., Sintek M., Biedert R., Ernst P., Dengel A. and Sonntag D..
REPRESENTING THE INTERNATIONAL CLASSIFICATION OF DISEASES VERSION 10 IN OWL.
DOI: 10.5220/0003082400500059
In Proceedings of the International Conference on Knowledge Engineering and Ontology Development (KEOD-2010), pages 50-59
ISBN: 978-989-8425-29-4
Copyright
c
2010 SCITEPRESS (Science and Technology Publications, Lda.)

(APHA) recommended that Canada, Mexico, and the
United States should also adopt it. Many other coun-
tries joined subsequently. Over the last 100 years it
was revised several times. The sixth revision included
morbidity and mortality conditions and was renamed
the “Manual of International Statistical Classification
of Diseases, Injuries and Causes of Death (ICD).”
Since 1948 the World Health Organization assumed
the responsibility for maintaining and publishing re-
vised versions of the ICD.
1
The currently internation-
ally effective revision is ICD-10 from 2006.
2
Although the overall structure of the ICD-10 was
accepted by numerous countries, different versions
exist which are maintained by national institutions.
For instance, the German version of the ICD-10 is
maintained by the DIMDI
3
which is under the author-
ity of the German Federal Ministry of Health. While
major parts of the ICD-10 hierarchy are equal both in
the DIMDI version and the WHO version, we found
out that the structure and content of certain parts of
the ICD-10 varies. Section 4 provides details of these
differences.
The aim of the work presented here is to gener-
ate an ontology covering the domain of human dis-
eases based on the classifications of the two country
specific ICD-10 versions described above. The ulti-
mate goal is to leverage technologies from the Se-
mantic Web to ease the work of medical experts by
supporting them in (1) making medical image data as
well as patient records available for semantic search,
and (2) by providing intelligent annotation sugges-
tions based on rich formal models for medical do-
main knowledge. This research was triggered by the
broader effort within the research project MEDICO.
From our discussions with clinicians we learned that
a representation of the ICD-10 is an absolute neces-
sity for efficient semantic radiological image annota-
tion in the everyday practice of the university hospital
participating in MEDICO (M
¨
oller et al., 2008).
2 RELATED WORK
The initial idea for generating an OWL version of
ICD-10 from data available on the web dates back to
a similar approach for generating an OWL model for
ICD-9 as presented in (M
¨
oller and Mukherjee, 2009).
1
“History of the development of the ICD,” available on
the WHO website at http://www.who.int/entity/classifica-
tions/icd/en/HistoryOfICD.pdf
2
http://www.who.int/classifications/icd/en/
3
Deutsches Institut f
¨
ur Medizinische Dokumentation
und Information
Biomedical ontologies and terminologies received
high attention in the last decade and provide promis-
ing technologies for data integration. Bodenreider
et al. evaluated popular large scale ontologies such
as SNOMED, FMA, and Gene Ontology and stated
that “ontologies play an important role in biomedi-
cal research through a variety of applications” (Bo-
denreider, 2004). In this context, a number of semi-
structured medical terminologies and classification
systems have been converted to formally structured
formats recently.
For the Systematized Nomenclature of Human
and Veterinary Medicine (SNOMED) (Cote et al.,
1993), an OWL ontology was created and used to
detect weaknesses in the original modeling (Schulz
et al., 2007; Schulz et al., 2009).
Noy and Rubin have presented an approach for
translating the Foundational Model of Anatomy on-
tology (FMA) to OWL (Noy and Rubin, 2008). From
their approach we adopted the idea to split the gen-
erated ontology into an OWL-DL and an OWL-Full
component.
Cardillo et al. presented an approach for a formal
representation of mappings between ICD-10 and the
International Classification of Primary Care version 2
(ICPC-2) (Cardillo et al., 2008). However, their fo-
cus was on the formal representation of mappings be-
tween ICD-10 and ICPC-2. The work presented in
this paper tries to complement their efforts by provid-
ing a formal model of additional relations within the
ICD-10.
3 APPROACH
This section describes our general approach for the
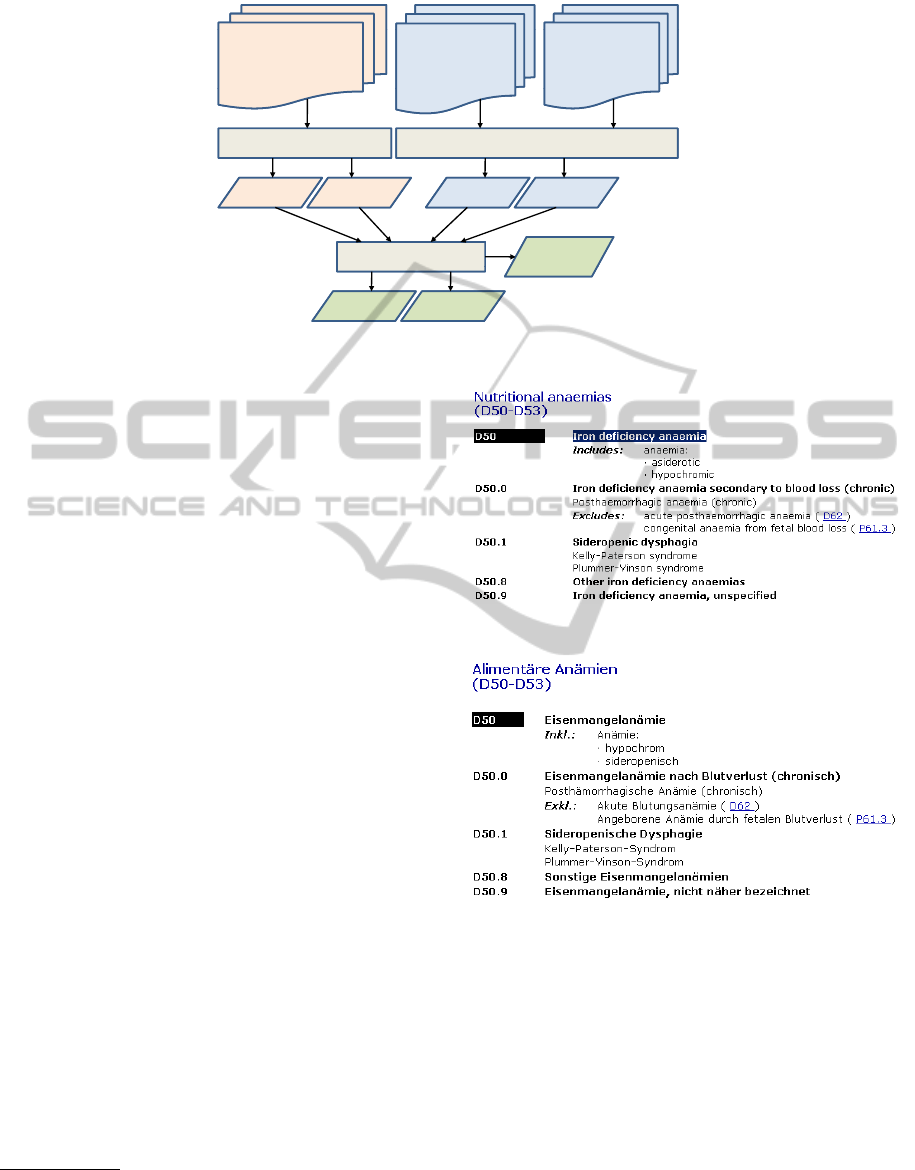
generation of the ICD-10 in OWL. Figure 1 shows
the data flow during the ontology generation process.
Following the elements in this diagram, the subse-
quent sections will discuss the different processing
steps and give details about the applied techniques
and algorithms.
3.1 Data Sources
The OWL ontology which we generated is based on
data available via the websites of the organizations
responsible for maintaining the respective ICD ver-
sions. As we will show, the data which is publicly
available on the Internet is well suited to generate a
rich formal model of the classification of human dis-
eases. The websites are highly structured and contain
enough information to fit the use case in the MEDICO
REPRESENTING THE INTERNATIONAL CLASSIFICATION OF DISEASES VERSION 10 IN OWL
51
GermanICDGermanICD‐‐1010
websitewebsite,,
maintainedmaintained byby
InternationalICDInternationalICD‐‐1010
websitewebsite,,maintainedmaintained
b
y
b
y
WHOWHO
GermanICDGermanICD‐‐1010
(XML(XMLfilefile))
DIMDIDIMDI
DIMDIDIMDI
yy
CrawlerCrawler
CrawlerCrawler
/XML Parser/XML Parser
CrawlerCrawler
CrawlerCrawler
/XML
Parser/XML
Parser
OWLOWL‐‐DLDL
componentcomponent
OWLOWL‐‐FullFull
componentcomponent
OWLOWL‐‐DLDL
componentcomponent
OWLOWL‐‐FullFull
componentcomponent
OWLOWL‐‐DLDL
componentcomponent
OWLOWL‐‐FullFull
componentcomponent
OWLOWL‐‐DLDL
componentcomponent
OWLOWL‐‐FullFull
componentcomponent
OntologyOntology MergerMerger
OWLOWL
differencesdifferences
filfil
OWLOWL‐‐DLDL
componentcomponent
OWLOWL‐‐FullFull
componentcomponent
OWLOWL‐‐DLDL
componentcomponent
OWLOWL‐‐FullFull
componentcomponent
fil
e
fil
e
Figure 1: Data flow of the ontology generation process.
project. Another advantage is that we can reflect up-
dates of the ICD published on the websites by re-
running our crawlers.
For the English version of the ICD we used the
official WHO website.
4
The website only partly re-
flects the original hierarchical structure of the ICD-
10. As an additional source we used the ICD-10
manual (WHO, 2004). Figure 2 (a) shows a screen-
shot covering the first of “Nutritional anaemias (D50-
D53).”
From the different German sources available we
chose to use the current ICD-10-GM, “GM” being the
“German Modification” (see Section 4). Figure 2 (b)
shows the same fragment of the ICD-10 as the previ-
ous screenshot, but this time in German. Our start-
ing points for the German ICD-10 is the respective
website and a publicly available XML file which is
structured using the Classification Markup Language
(ClaML). As the name suggests, ClaML is special
language designed to represent classification hierar-
chies (Hoelzer et al., 2002). It provides special no-
tations to state super- and subclass relations, declare
attributes, and to specify metadata elements, among
other things. To interpret the notation correctly,
we use the WHO manual (WHO, 2004) and a sup-
plementary documentation for attributes exclusively
stated in the DIMDI version (Deutsche Krankenhaus-
gesellschaft, 2009) using the the DIMDI website.
5
3.2 OWL Model Generation
The general structure of the ICD-10 is as follows. It
consists of “Chapters” using Roman numerals from I
4
http://apps.who.int/classifications/apps/icd/
icd10online/
5
http://www.dimdi.de/static/de/klassi/diagnosen/icd10/
htmlgm2009/index.htm
(a) Example for an English entry from the WHO ICD-10
website
(b) Respective entry from the German DIMDI ICD-10 web-
site
Figure 2: Language-specific ICD-10 online versions.
to XXI. The chapters again contain “Blocks of cat-
egories” (e. g., Chapter III: “Diseases of the blood
and blood-forming organs and certain disorders in-
volving the immune mechanism”) which specify a
range of categories of a particular aspect (e. g., D50-
D89). These blocks then contain “Categories,” de-
noted by an ICD-10 code, a capital letter, and Ara-
bic numbers (e. g., D50-D53: Nutritional anaemias;
D55-D59: Haemolytic anaemias; etc.). They are
further subdivided into “Subcategories.” The sub-
KEOD 2010 - International Conference on Knowledge Engineering and Ontology Development
52

categories are coded by attaching an additional digit
after the decimal point (e. g., D50.1: Sideropenic
dysphagia) . These codes differentiate from the
specific language or writing system of the differ-
ent ICD-10 versions. Contrary to the WHO web-
site, the DIMDI XML file constrains the subcate-
gories by using an additional decimal number, which
is appended to the codes of their parent categories.
These subcategories are also defined using “Modi-
fiers” which specify a set of “ModifierClasses” as
subclasses. Each of these classes possesses a number,
a label, and additional information such as “Exclu-
sions” or “Inclusions.” If a category contains a “Mod-
ifier,” it will be specialized by generating new cate-
gories with each particular “ModifierClass.” These
relations are represented in OWL by creating a new
OWL class for each “Modifier,” which is a subclass of
icd10:Modifier, and defining the appropriate Mod-
ifierClasses as subclasses. The combination is de-
noted using an owl:unionOf of the specific category
and each ModifierClass. This form of specification is
used extensively. Our analysis has shown that 4488 of
the 16214 classes are specified in this way by DIMDI.
ICD-10 is not only a classification of diseases, but
the terminology also includes links to other related
aspects, such as symptoms, signs and consequences
of other external causes. Therefore, the manual de-
scribes an additional level of order which groups cer-
tain chapters according to their particular aspects. For
example, “Chapters I to XVII relate to diseases and
other morbid conditions.” It is worth mentioning that
this systematic level is not available from the website
but only from the manual.
Using this information about chapters and groups
of chapters we modeled the first two hierarchy levels
by hand. The OWL class icd10:Entry is the super
class of the bilingual ICD-10 hierarchy. As mentioned
before, differences between the German and English
ICD-10 exist. Our analysis shows that there are ICD-
10 categories which are present in the German ICD-
10 but not in the English ICD-10 and vice versa. Sec-
tion 4 gives details about these differences.
In addition, the origins of the concepts are also
encoded in our OWL model. The first approach was
to add a super class for each class to denote its ori-
gin. But this proved to be incorrect. Let us con-
sider the block R00-R99, which is present in both
terminologies and thus gets both super classes. The
symptom R65, which has the super class R00-R99, is
only stated by the DIMDI. Consequently, a reasoner
would infer that R65 is in the DIMDI and WHO ver-
sion, because it would build the transitive closure and
R65 would get both super classes. For that reason,
we decided to denote the provenance using the two
Figure 3: General class hierarchy of the OWL model.
boolean OWL-Full properties icd10:isDIMDIEntry
and icd10:isWHOEntry.
Figure 3 shows an (abbreviated) example of
the generated class hierarchy. We use two HTTP
crawlers, implemented in Java, to generate OWL
models for each of the two input sources. OWL
classes and axioms are generated using the Jena On-
tology API.
6
Other libraries—such as the OWL API
(Bechhofer et al., 2003)—were not able to handle the
OWL-Full expressivity of our modeling.
The generated OWL model consists of two com-
ponents. The OWL-DL part contains the hierarchy
of the ICD-10 according to the hierarchical structure
described above. All ICD-10 categories and subcate-
gories are reflected by OWL classes. The hierarchical
information is reflected by owl:subClassOf axioms.
For a discussion of the contents of the OWL-Full part
see further below.
We will explain the next steps by giving an exam-
ple. Figure 2 shows the first part of the WHO ICD-
10 website about “Nutritional anaemias (D50-D53).”
We will focus on the entry “D50.0 Iron deficiency
anaemia secondary to blood loss (chronic)” as a guid-
ing example throughout this paper.
Each class is identified by an URL, which con-
sists of a specific ICD-10 name space and the special
term as the URL anchor. The terms for the categories
are simply the particular ICD-10 codes. For blocks
6
http://jena.sourceforge.net/ontology/
REPRESENTING THE INTERNATIONAL CLASSIFICATION OF DISEASES VERSION 10 IN OWL
53

and chapters, a range pattern is used which covers
their content, e. g., D50-D53. From this we create an
OWL class with the local name “D50.0.” The “.0”
indicates that this is a sub-category of “D50 Iron defi-
ciency anaemia.” Thus, we add an owl:subClassOf
axiom which represents this relationship. The bold-
faced name of the sub-category becomes the En-
glish rdfs:label of this class. Later, by merging
with the OWL model of the German ICD-10, we
can also add the German labels “Eisenmangelan
¨
amie
nach Blutverlust (chronisch).” For some concepts,
the DIMDI specifies up to three labels, which dif-
fer in their length and detail. The smaller labels are,
thereby, necessary because some print formats require
them. In our case we can neglect this limitation and
use always the most detailed label available. We use
the standard XML language tags to differentiate be-
tween these languages.
3.3 ICD-10 Characteristics
Despite a specialization hierarchy, multiple character-
istics are stated in the WHO manual (WHO, 2004)
and the DIMDI supplement (Deutsche Krankenhaus-
gesellschaft, 2009), and they can be shared by differ-
ent classes. These are:
Dagger and Asterisk Categories. Statements con-
taining information about an underlying disease
with a particular additional manifestation can be
expressed thanks to asterisk and dagger codes.
Underlying diseases are marked with a dagger and
are the primary criterion. Therefore, they have
to appear in the diagnostic statement, whereas
the manifestation marked with an asterisk is
only additional. These circumstances are rep-
resented in OWL using two properties, namely
icd10:hasAdditionalManifestation with its
inverse icd10:hasUnderlyingDisease. The
first one’s domain is all classes which represent
a dagger category and have a range of all asterisk
categories. The restriction that an additional man-
ifestation needs at least one underlying disease is
expressed by the property’s cardinality, which is
at least one.
Optional Concepts. The DIMDI defined a supple-
mental characteristic and this is only applied in
their version of the terminology. Optional con-
cepts are similar to dagger and asterisk categories.
If marked as optional, a concept will be manda-
tory for some diagnoses but only supplemental for
other ones.
Categories Limited to one Gender. The ICD-10
contains several categories which are only ap-
plicable to either males or females. Consider,
for instance, diseases of the genitals, like “D40
Neoplasm of uncertain or unknown behavior of
male genital organs” or conditions which occur
during the pregnancy of women, e. g., “O00
Ectopic pregnancy.” The facts are represented by
one super class for each gender.
Sequelae Categories. Sequelae categories are used
for mortality cause encoding. They indicate that
the death is not caused by the main effect of a
given disease. Instead it is caused by residual ef-
fects.
Postprocedural Disorders. Categories which fall
under this characteristic point out conditions and
complications which occur after treatment, e. g.,
surgical wound infections or shock.
Contrary to dagger and asterisk categories, each char-
acteristic is represented using a specific super class.
All classes which share the characteristic are sub-
classes of this class.
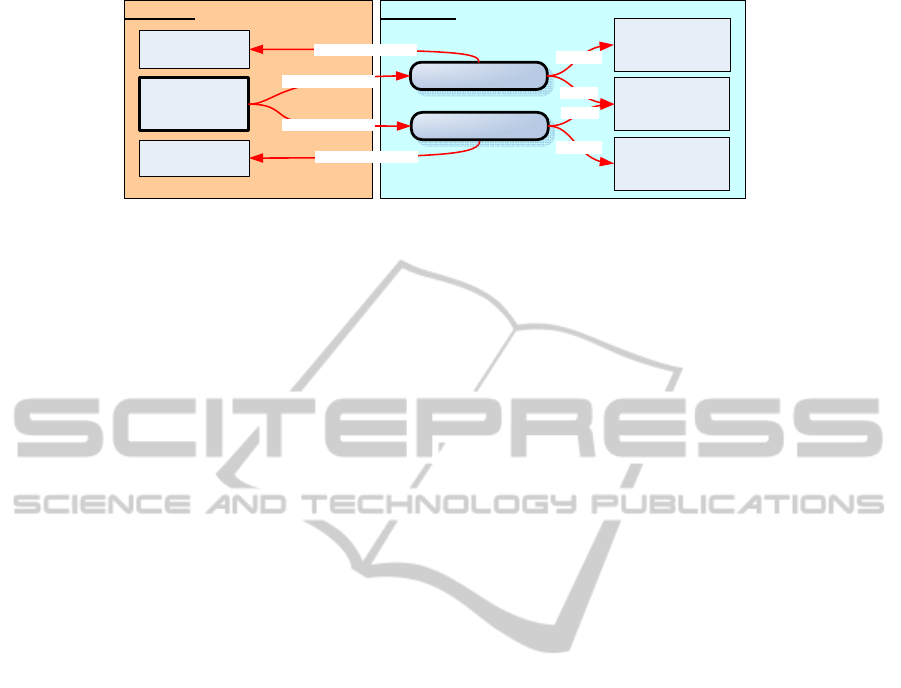
3.4 Handling ICD-10 Exclusions
For some ICD-10 categories so-called “Exclusions”
also exist. According to the ICD-10 manual (WHO,
2004), they exclude certain conditions that, “although
the rubric title might suggest that they were to be clas-
sified there, are in fact classified elsewhere.” The
example in Figure 2 (a) lists two such excludes for
D50.0: “acute posthaemorrhagic anaemia” with a link
to ICD-10 category D62 and “congenital anaemia
from fetal blood loss” with a link to category P61.3.
We capture this information by adding owl:dis-
jointWith axioms between D50.0 and D62 as well
as between D50.0 and P61.3. This can be expressed
using the expressivity of OWL-DL (see Figure 4).
However, by relying exclusively on owl:dis-
jointWith axioms, we would lose important infor-
mation. As the ICD-10 manual states, exclusions can
be extended using additional strings and constructions
of braces. They indicate that neither the words that
precede them nor the words after them are proper
terms. Thus, a more precise qualification has to be
applied (WHO, 2004). If we compare the brace con-
structs with the encoding in the XML file, it becomes
clear that they are used to provide a more compre-
hensive structuring of the data. The XML file reflects
them by splitting up the “Exclusions” and adding a
new fragment for each brace element to the “Ex-
clusions.” Figure 5 depicts “Inclusions” for concept
“O71.6: Obstetric damage to pelvic joints and lig-
aments” using braces and figure 6 shows them us-
ing the German XML encoding. As we see, without
proper post-processing we are not able to relate that
KEOD 2010 - International Conference on Knowledge Engineering and Ontology Development
54
OWL-FullOWL-DL
icd10:ICDDescription
rdf:type
icd10:hasExcludes
icd10:concernsClass
icd10:concernsClass
congenital anaemia
from fetal blood loss
rdfs:label
icd10:P61.3
icd10:D62
icd10:D50.0
icd10:
ICDDescription
icd10:ICDDescription
acute
posthaemorrhagic
anaemia
rdfs:label
icd10:hasExcludes
rdf:type
Figure 4: Structure and relationship of OWL-DL and OWL-Full component by example.
the “Exclusion” of concept M54.1 is shared by multi-
ple Exclusions.
These qualifications are covered in additional
OWL individuals of class icd10:Description. The
individuals can have several properties of type
icd10:concernsClasses. Because this property
concerns other ICD-10 categories, it needs to have
a class-valued range. Thus, the individuals require
OWL-Full expressivity. To encode the string infor-
mation of an “Exclusion,” we use rdfs:label. For
each excluded statement, we generate one individ-
ual. We also encode the information contained in the
brace constructs, which appear in the WHO version.
Therefore, we create an individual for the information
which occurs after the braces. They are related to each
Exclusion using an OWL-Full property icd10:qua-
lifiedBy. Extracting this information for the DIMDI
data is among our next steps.
Additionally, a closer look at the ICD-10 revealed
that for numerous categories, the “Exclusions” do
not point to other ICD-10 categories but to arbi-
trary descriptions of certain medical symptoms. Fig-
ure 5 gives an example showing ICD-10 subcategory
“O71.6: Obstetric damage to pelvic joints and liga-
ments.” As we cannot generate proper disjointness
axioms for these exclude expressions we decided to
store them using the exclude individuals described
above without pointing to another ICD-10 category.
3.5 Handling ICD-10 Inclusions and
Notes
Similar to the “Exclusions” two other properties for
categories are part of the ICD-10. “Inclusions” are ad-
ditions to the rubric in which they occur. The ICD-10
manual describes them as a guide and provides exam-
ples to formulate diagnostic statements. “Inclusions”
are represented using the individuals in OWL-Full in
the same way as “Exclusions.”
In addition, it is possible that a note is provided
for an ICD-10 element. These notes give hints how
to use the particular category, block, or chapter. For
example, a physician who is writing a medical report
sees from these hints that the category “G09 Seque-
lae of inflammatory diseases of central nervous sys-
tem” is to be used to indicate conditions whose pri-
mary classification is G00-G08 (i. e., excluding those
marked with an asterisk) as the cause of sequelae,
themselves classifiable elsewhere. The only purpose
of the notes is to support human beings in interpret-
ing the ICD-10. Also, they are not interpretable for
reasoners because they only contain continuous text.
For that reason, we do not relate the notes individuals
to classes with OWL-Full properties, instead we use
owl:AnnotationProperties.
3.6 Merging the English and German
OWL Models
To merge the two ICD-10 variants, we have to distin-
guish between the OWL-DL and -Full parts. The two
OWL-Full parts are merged by just importing them
into a new ontology. This is possible because they
only contain properties of the classes defined in the
DL versions and the class definitions were not altered
during the merging process.
The merging of the DL ontologies can be divided
into two phases. First, an automatic integration is per-
formed, which is then refined by a manual step. The
automatic merging process starts with the WHO on-
tology. As stated in the last paragraph of section 3.2,
all classes are identified using their particular ICD-10
code. In most of the cases, these codes are the same
in the DIMDI and WHO version. Therefore, we be-
gin the integration by checking if each DIMDI class
is present in the WHO version. If so, we add the label
and owl:subClassOf properties of the class. Adding
owl:subClassOf axioms is necessary because only
some of them exist in the DIMDI version. For ex-
ample, if a block only appears in the DIMDI version,
all owl:subClassOf relations concerning this block
only occur in the DIMDI ontology. If the class is not
contained, it will be created and all properties will be
copied.
REPRESENTING THE INTERNATIONAL CLASSIFICATION OF DISEASES VERSION 10 IN OWL
55
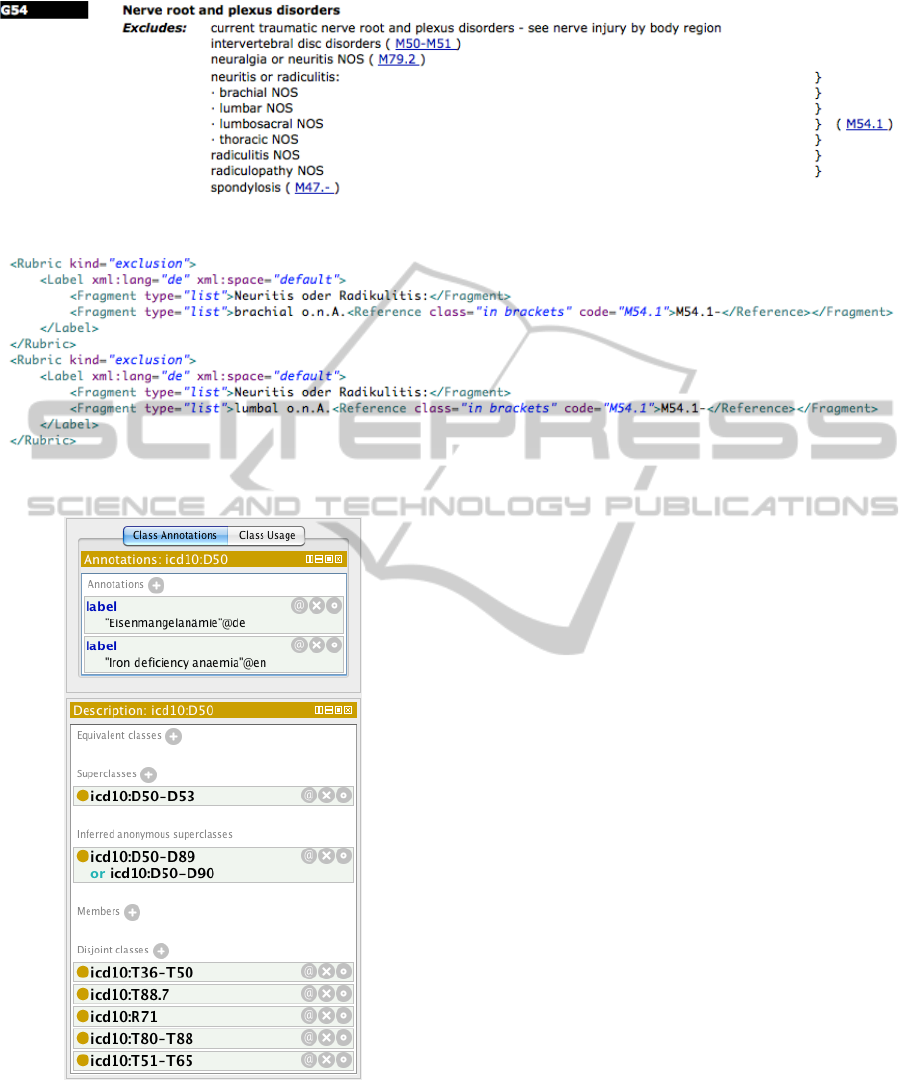
Figure 5: Example for ICD-10 category with “Exclusions” that do not point to other ICD-10 categories.
Figure 6: Example for ICD-10-GM braces constructs within “Exclusions”.
Figure 7: Screenshot of the OWL-DL version of the ICD-10
in the Ontology Editor Protg.
In addition, a few classes exist which are seman-
tically very similar, but differ by their ICD code. For
example, the DIMDI chapter D50-D90 only varies in
the range and the existence of the class D90 from the
WHO chapter D50-D89, but both concern the same
diseases. To merge these classes, we first manually
determine all possible pairs (classA, classB) which
differ in this sense. After that, we define a new super
class unionAB for each pair. This is the owl:unionOf
of the pair’s classes and gets the a concatenation of
the local names of both. To determine the location of
the new class, we search the first super class which
classA and classB have in common. unionAB is then
added as a subclass of this class. This traversing is
necessary, because there can be super classes not cov-
ering the entire range of unionAB. For example, we
merge the blocks D80-D89 and D80-D90 and the di-
rect super classes are D50-89 and D50-D90 respec-
tively, which are only present either in the WHO or
DIMDI version. Therefore, we have to traverse the hi-
erarchy one step further and find the appropriate super
class, which is owl:unionOf of the classes D50-D89
and D50-D90.
Besides the new integrated ontology, a differ-
ence ontology is generated during the merging pro-
cess, which distinguishes between WHO and DIMDI
classes only occurring in one version. We know
that this knowledge is already contained in the on-
tology by the label and icd10:isDIMDIEntry or
icd10:isWHOEntry. However, we decided to pro-
duce an explicit representation of the differences, be-
cause it makes the merging process more transparent
and the differences are easier to examine. For these
reasons, we denote the differences in both ontologies
using OWL-Full properties in the difference ontology.
We are using one property for every ICD source to de-
KEOD 2010 - International Conference on Knowledge Engineering and Ontology Development
56

note the exclusiveness and one property to denote the
classes later manually added by the merging process.
Figure 7 shows a screenshot of the generated OWL
class for ICD-10 category D50.0 in Protg
7
.
4 RESULTS AND DISCUSSION
By the definition given in the ICD-10 manual, the ICD
is a classification system with “a hierarchical structure
with subdivisions.” And further: “A statistical classi-
fication of diseases should retain the ability both to
identify specific disease entities and to allow statis-
tical presentation of data for broader groups, to en-
able useful and understandable information to be ob-
tained.” From this we concluded that the ICD is based
on a hierarchical system of classes. The relations be-
tween these classes are proper subset relations in the
sense of set theory. Thus, we decided to represent the
relations of the ICD using OWL and its subClassOf
relations.
Table 1 lists some general metrics for the gener-
ated ontology. It also lists differences between the
German and the English ICD-10 versions in terms of
number of classes. The majority of all ICD-10 cate-
gories, i. e., about 60%, share the same ICD-10 code
(for details see Section 3.2) and thus could be mapped
using this as an identifier. However, there were some
discrepancies between the WHO and German ver-
sions. One reason were different modeling granu-
larities producing more categories in some branches.
These differences are discussed subsequently.
We decided to split our OWL model into two
components similar to the approach of Noy and Ru-
bin in (Noy and Rubin, 2008) for translating the
Foundational Model of Anatomy ontology to OWL.
The OWL-DL component allows to perform DL-
reasoning using standard OWL-DL reasoners like
Pellet (Sirin et al., 2007). Information from the ICD
which requires modeling in OWL-Full is still avail-
able in the OWL-Full component. To use the com-
plete model, the OWL-Full component can be loaded.
This variant imports the OWL-DL model.
Differences between WHO and DIMDI
Versions of ICD-10
In both terminologies we located classes that either
only occur in the DIMDI or WHO version. These
differences can be classified into two categories:
• classes which only appear in one ICD-10 variant
and have no particular counterpart in the other and
7
http://protege.stanford.edu
• classes which have a slightly different identifica-
tion, but can be merged manually.
The first category is the most extensive. We iden-
tified 1,145 classes which are exclusively part of the
WHO version and 5,707 classes exclusively part of
the DIMDI version. It is important to note that we
are only regarding the classes of the actual ICD-10
entries and not the classes for constructs like mod-
ifiers; these will be discussed later. Furthermore,
there are blocks which differ in both versions. This
means that one version specifies some parts of its
terminology with more granularity than the other or
that some concepts were simply left out. For exam-
ple, the WHO subdivides the block “V01-X59 Ac-
cidents” into 27 sub-blocks using two hierarchy lev-
els. In contrast, the DIMDI version does not make
any further subdivisions here. Moreover, the DIMDI
describes the block “U60-U61 Stadieneinteilung der
HIV-Infektion” (“Staging of HIV-Infection”), which
is not present in the WHO version at all.
We derived the second category during a manual
examination of all differences. Hereby, we identified
five blocks, which vary in their ICD-10 code. Ta-
ble 2 lists them and opposes the WHO blocks with
the details which the DIMDI contains. In all cases,
there are differences in the range of the blocks. This
is interesting, as even though a block can specify a
broader range, it can be semantically more restricted.
We will illustrate this by an example. The WHO ver-
sion has the block “U80-U89 Bacterial agents resis-
tant to antibiotics.” The German version “U80-U85
Infektionserreger mit Resistenzen gegen bestimmte
Antibiotika oder Chemotherapeutika” has almost the
same range (“U80-U89” vs. “U80-85”). It could be
assumed that the block with the smaller range is also
more specific. But in this example the opposite is true
since the German term “Infektionserreger” (“infec-
tious agent”) includes diseases caused by both bacte-
ria and viruses. In contrast, the WHO block only cov-
ers diseases caused by bacteria. Section 3.6 describes
our manual approach for merging such classes.
5 CONCLUSIONS AND FUTURE
WORK
In this paper we presented our approach for modeling
the hierarchy of the ICD-10 in OWL. Our model cap-
tures the hierarchical information of ICD-10 as well
as comprehensive class labels both for English and
German. Peculiarities such as “Exclusions,” state-
ments which make statements about the disjointness
of certain ICD-10 categories, are provided in a sepa-
rate OWL-Full component. This component allows
REPRESENTING THE INTERNATIONAL CLASSIFICATION OF DISEASES VERSION 10 IN OWL
57

Table 1: Metrics for the generated ICD-10 ontology.
WHO ICD-10 German ICD-10
OWL classes 11,308 16,214
disjointness axioms (see Section 3.4) 13,094 27,899
excludes pointing to another category 5,150 4,417
excludes without a proper link to other categories 35 73
Table 2: ICD-10 blocks, which can be merged together, although they exclusively appear in the DIMDI or WHO version.
WHO ICD-10 DIMDI ICD-10
D50-D89 Diseases of the blood and blood-forming
organs and certain disorders involving the immune
mechanism
D50-D90 Krankheiten des Blutes und der blutbilden-
den Organe sowie bestimmte Strungen mit Beteili-
gung des Immunsystems
D80-D89 Certain disorders involving the immune
mechanism
D80-D90 Bestimmte Strungen mit Beteiligung des
Immunsystems
V01-Y98 External causes of morbidity and mortality V01-Y84
¨
Auere Ursachen von Morbiditt und Mor-
talitt
O80-O84 Delivery O80-O82 Entbindung
U80-U89 Bacterial agents resistant to antibiotics U80-U85 Infektionserreger mit Resistenzen gegen
bestimmte Antibiotika oder Chemotherapeutika
the use of hierarchical knowledge and class labels
with existing OWL-DL reasoners. Our automatic
generation and merging method also revealed system-
atic differences between the German DIMDI and the
English WHO version.
The goal of this approach was to partly reduce
Semantic Heterogeneity in health care, mentioned in
Section 1 by integrating two semi-formal terminolo-
gies. We plan to combine the results with addi-
tional conceptualizations next, so that ontology net-
works can be created and interconnected. For ex-
ample, Cardillo et al. describe an approach to map
the ICD with the International Classification of Pri-
mary Care Version 2 (ICPC-2) (Cardillo et al., 2008).
This would foster the creation of expressive medical
knowledge bases which would improve the retrieval
and reuse of knowledge by reducing ambiguity.
The generated OWL ontology is used in the re-
search project MEDICO to allow for semantic anno-
tation and retrieval across medical documents and im-
ages annotated with ICD-10 terms both in English and
German.
The current ontology represents the main and
most important parts of both ICD-10 variants. How-
ever, the XML file states additional information to
be integrated in the future. For example, the dag-
ger and asterisks are distinguished depending on the
treatment. Different asterisk and dagger terms have
to be used to formulate diagnoses for clinical treat-
ments if diagnosis reports are created for accounting
documents. During our examination of the generated
OWL files and the respective ICD-10 websites, we
recognized that certain verbal structures occur very
often, e. g., “Injury of X,” where X is an anatomi-
cal designation, like arm or leg. Moreover, the man-
uals describe predefined terms which are used very
often, for example, the two acronyms NOS, meaning
“not otherwise specified,” and NEC, standing for “not
elsewhere classified.” Linguistic analysis can exploit
this information to extract relations, e. g., to concepts
represented in anatomical ontologies. This approach
has already been applied successfully to other corpora
within the MEDICO project (Wennerberg et al., 2009;
Wennerberg, 2009) and we plan to extend it to the
ICD-10 as well.
Another possibility to enhance our OWL models
is to include more languages. This will generate an
international representation of the ICD-10. For exam-
ple, it would be possible to include the French ver-
sion of the ICD-10
8
. The website is structured like
the German DIMDI version. Consequently, either our
HTML crawler could be used to extract the necessary
information or–if a XML encoding is available–this
could be parsed directly. However, at first appearance
one can see that the French version also differs from
the other two version, e. g., it covers only 21 chapters
8
http://www.dimdi.de/dynamic/en/klassi/diagnosen/
icd10/htmlfren/fr-icd.htm
KEOD 2010 - International Conference on Knowledge Engineering and Ontology Development
58

and omits the chapter about “Codes for special pur-
poses.” Therefore, an examination of the differences
will be necessary.
ACKNOWLEDGEMENTS
This research has been supported in part by the THE-
SEUS Program in the MEDICO Project, which is
funded by the German Federal Ministry of Economics
and Technology under grant number 01MQ07016.
The responsibility for this publication lies with the au-
thors.
REFERENCES
Bechhofer, S., Volz, R., and Lord, P. W. (2003). Cooking
the semantic web with the OWL API. In International
Semantic Web Conference, pages 659–675.
Bodenreider, O. (2004). The Unified Medical Language
System (UMLS): Integrating biomedical terminology.
Nucleic Acids Research, 32 (Database Issue):D267–
D270.
Cardillo, E., Eccher, C., Serafini, L., and Tamilin, A.
(2008). Logical analysis of mappings between med-
ical classification systems. In Dochev, D., Pistore,
M., and Traverso, P., editors, AIMSA, volume 5253 of
Lecture Notes in Computer Science, pages 311–321.
Springer.
Cote, R., Rothwell, D., Palotay, J., Beckett, R., and Brochu,
L. (1993). The systematized nomenclature of human
and veterinary medicine. Technical report, SNOMED
International, Northfield, IL: College of American
Pathologists.
Deutsche Krankenhausgesellschaft (2009). Deutsche
kodierrichtlinien - allgemeine und spezielle kodier-
richtlinien f
¨
ur die verschl
¨
usselung von krankheiten
und prozeduren. Technical report, Institut f
¨
ur das Ent-
geltsystem im Krankenhaus (InEK GmbH).
Hoelzer, S., Schweiger, R. K., Liu, R., Rudolf, D., Rieger,
J., and Dudeck, J. (2002). Xml representation of hier-
archical classification systems: from conceptual mod-
els to real applications. Proc AMIA Symp, pages 330–
4.
Lenz, R. (2005). Information management in distributed
healthcare networks. Data Management In a Con-
nected World, 3551:315–334.
M
¨
oller, M. and Mukherjee, S. (2009). Context-Driven On-
tological Annotations in DICOM Images – Towards
Semantic PACS. In Azevedo, L. and Londral, A. R.,
editors, Proceedings of the Second International Con-
ference on Health Informatics, HEALTHINF, pages
294–299. INSTICC Press.
M
¨
oller, M., Sintek, M., Buitelaar, P., Mukherjee, S., Zhou,
X. S., and Freund, J. (2008). Medical image under-
standing through the integration of cross-modal object
recognition with formal domain knowledge. In Proc.
of HEALTHINF 2008, volume 1, pages 134–141, Fun-
chal, Madeira, Portugal.
Noy, N. F. and Rubin, D. L. (2008). Translating the Founda-
tional Model of Anatomy into OWL. Web Semantics:
Science, Services and Agents on the World Wide Web,
6(2):133–136.
Schulz, S., Suntisrivaraporn, B., and Baader, F. (2007).
SNOMED CT’s problem list: Ontologists’ and logi-
cians’ therapy suggestions. In Proc. of The Medinfo
2007 Congress, Studies in Health Technology and In-
formatics (SHTI-series). IOS Press.
Schulz, S., Suntisrivaraporn, B., Baader, F., and Boeker, M.
(2009). SNOMED reaching its adolescence: Ontolo-
gists’ and logicians’ health check. International Jour-
nal of Medical Informatics, 78(Supplement 1):S86–
S94.
Sirin, E., Parsia, B., Grau, B., Kalyanpur, A., and Katz, Y.
(2007). Pellet: A practical owl-dl reasoner. Web Se-
mantics: Science, Services and Agents on the World
Wide Web, 5(2):51–53.
Sonntag, D. (2010). Ontologies and Adaptivity in Dialogue
for Question Answering. AKA and IOS Press, Heidel-
berg.
Sonntag, D., Wennerberg, P., Buitelaar, P., and Zillner, S.
(2009). Pillars of ontology treatment in the medical
domain. Journal of Cases on Information Technology
(JCIT), 11(4):47–73.
Wennerberg, P. (2009). Aligning medical domain ontolo-
gies for clinical query extraction. In Proc. of the 12th
Conference of the European Chapter of the Associa-
tion for Computational Linguistics: Student Research
Workshop (EACL ’09), pages 79–87, Morristown, NJ,
USA. Association for Computational Linguistics.
Wennerberg, P., M
¨
oller, M., and Zillner, S. (2009). A lin-
guistic approach to aligning representations of human
anatomy and radiology. In Proc. of the International
Conference on Biomedical Ontologies (ICBO 2009).
WHO (2004). International statistical classification of dis-
eases and related health problems. Technical report,
World Health Organization.
REPRESENTING THE INTERNATIONAL CLASSIFICATION OF DISEASES VERSION 10 IN OWL
59
