
GENERATING LITERATURE-BASED KNOWLEDGE
DISCOVERIES IN LIFE SCIENCES USING
RELATIONSHIP ASSOCIATIONS
Steven B. Kraines, Weisen Guo, Daisuke Hoshiyama, Haruo Mizutani
Science Integration Program (Human), Department of Frontier Sciences and Science Integration
Division of Project Coordination, The University of Tokyo, 5-1-5 Kashiwa-no-ha, Kashiwa, Chiba, 277-8568, Japan
Toshihisa Takagi
Department of Bioinformatics, School of Frontier Science, The University of Tokyo
5-1-5 Kashiwa-no-ha, Kashiwa, Chiba, 277-8568, Japan
Keywords: Relationship associations, Association rules, Semantic relationships, Semantic matching, Semantic web,
Ontology, Logical inference, Life sciences, Literature-based knowledge discovery.
Abstract: The life sciences have been a pioneering discipline for the field of knowledge discovery, since the literature-
based discoveries by Swanson three decades ago. Existing literature-based knowledge discovery techniques
generally try to discover hitherto unknown associations of domain concepts based on associations that can
be established from the literature. However, scientific facts are more often expressed as specific relation-
ships between concepts and/or entities that have been established through scientific research. A pair of rela-
tionships that predicate the specific way in which one concept relates to another can be associated if one of
the concepts from each relationship can be determined to be semantically equivalent; we call this a “rela-
tionship association”. Then, by making the same assumption of the transitivity of association used by Swan-
son and others, we can generate a hypothetical relationship association by combining two relationship asso-
ciations that have been extracted from a knowledge base. Here we describe an algorithm for generating po-
tential knowledge discoveries in the form of new relationship associations that are implied but not actually
stated, and we test the algorithm against a corpus of almost 5000 relationship associations that we have ex-
tracted in previous work from 392 semantic graphs representing research articles from MEDLINE.
1 INTRODUCTION
In the 1980’s, Don Swanson demonstrated that bona
fide scientific discoveries can be made just by exam-
ining the co-occurrence of scientific concepts in
research articles that already exist in the literature
(Swanson, 1986). He noted that several research
articles mentioned “Raynaud's syndrome”, which
results in discoloration of extremities, together with
intermediary concepts such as “blood viscosity”.
Other articles mentioned the same intermediary con-
cepts together with “fish oil”. However, no articles
mentioned “fish oil” and “Raynaud’s syndrome”
together. This led him to hypothesize that fish oil is
effective for treating Raynaud’s syndrome, a scien-
tific discovery that was later experimentally verified.
Swanson made several other discoveries from the
literature using this technique, which became known
as the Swanson ABC model (Swanson, 1990).
Swanson’s discoveries gave birth to the field of “lit-
erature-based knowledge discovery” and led to a
widespread belief in the information science com-
munity that not only could discoveries be made from
the existing literature, but those discoveries could be
made entirely automatically. Several attempts to
develop computational methods that can automati-
cally discover new scientific knowledge or generate
novel hypotheses from the existing literature have
been reported (Langley, 2000; Racunas et al., 2004,
Srinivasan, 2004, Weeber et al., 2005). However,
there have been few reports of actual new discover-
ies made from the literature since the initial discov-
eries by Swanson (Natarajan et al. (2006) reported
35
Kraines S., Guo W., Hoshiyama D., Mizutani H. and Takagi T..
GENERATING LITERATURE-BASED KNOWLEDGE DISCOVERIES IN LIFE SCIENCES USING RELATIONSHIP ASSOCIATIONS .
DOI: 10.5220/0003068100350044
In Proceedings of the International Conference on Knowledge Discovery and Information Retrieval (KDIR-2010), pages 35-44
ISBN: 978-989-8425-28-7
Copyright
c
2010 SCITEPRESS (Science and Technology Publications, Lda.)

one discovery that was made in part through exami-
nation of the literature).
In a clever article entitled “In silico veritas”, Al-
len criticized a predominant attitude in the scientific
community that “computers can do our thinking for
us” (Allen, 2001). In a follow-up response, Smal-
heiser, who worked with Swanson on automating
some parts of the ABC model (Swanson and Smal-
heiser, 1997), made it clear that the literature-based
knowledge discovery techniques “do not attempt to
bypass scientists, but rather help them to integrate
knowledge that is retrievable from the scientific lit-
erature in order to formulate hypotheses quickly,
systematically and comprehensively.” He went on to
say that the process would be even more effective “if
investigators and funding agencies simply included
archiving of samples and data into research projects
together with the metadata needed to understand
how the data were collected” (Smalheiser, 2002). In
other words, although Swanson was able to make
interesting scientific discoveries just by examining
the standard research deliverables that had been cre-
ated by researchers in human-readable form, if re-
searchers and disseminators of scientific knowledge
were to present scientific knowledge in a form that is
directly interpretable by computers, the benefits to
increasing the effectiveness of in silico methods for
scientific discovery would be considerably larger.
The idea of getting the scientific community to
create computer-readable descriptors of their re-
search articles, such as structured digital abstracts,
has been brought up recently (Gerstein et al., 2007;
Ceol et al., 2008; Seringhaus and Gerstein, 2008).
The proposed structures for the descriptors make the
content of research articles more accessible to search
engines, text mining systems and perhaps even hu-
man readers (Hartley and Betts, 2007). However,
even in structured digital abstracts, the granularity of
“cognition” for most of the descriptive information
is still at the sentence or paragraph level (Ceol et al.,
2008). Consequently, computers still need to make
sense of the sentences in the delimited entries in the
digital abstracts (Cafarella et al., 2007; O'donnell et
al. 2001), which is notoriously difficult due to the
complexity and ambiguity of natural language (Na-
tarajan et al., 2005; Hunter & Cohen, 2006).
Our aim is to take the idea of creating computer-
readable content in the scientific knowledge dis-
semination process one step further. Specifically, we
hypothesize that by drawing on new techniques and
standards for semantic representation of knowledge
in a computer-interpretable form, it should be possi-
ble for human researchers to create descriptors of
their research findings that are not just “computer-
readable” but also “computer-understandable”. By
“computer-understandable”, we mean that com-
puters can reason with the semantics of the descrip-
tors in reference to shared mental models or concep-
tualizations of the knowledge domain and that they
can infer new “facts” or “assertions” in the form of
relationships between concepts and/or entities that
are only implied but not explicitly stated.
Here, we present an algorithm for discovering
hypotheses based on associations between specific
relationships, called “relationship associations”. The
relationship associations are mined from computer-
understandable descriptors in the form of semantic
graphs. In order to demonstrate the potential effec-
tiveness of this approach, we apply the algorithm to
a corpus of semantic graphs that we have created
previously. We then describe some of the hypotheti-
cal relationship associations that are discovered.
This paper is organized as follows. In Section 2,
we present previous work that forms the background
of our study. In Section 3, we describe our algorithm
for generating hypothetical relationship associations
that represent new and potentially meaningful asso-
ciations of specific relationships. In Section 4, we
report the results of an experiment applying this al-
gorithm to the corpus of semantic graphs created
previously. In Section 5, we review related work.
2 BACKGROUND
Current text mining techniques cannot accurately
extract semantic relationships between concepts
from natural language text due to the complexity and
ambiguity of natural language (Erhardt et al., 2006;
Rinaldi et al. 2006). We have developed a system
that uses ontologies based on Description Logics
(DL) to enable researchers to author semantic graphs
that define the relationships described by a research
article in a computer understandable form (Kraines
et al., 2006). By using DL ontologies as formal
knowledge representation languages for authoring
the semantic graphs, it is possible to accurately ex-
press specific relationships between concepts in a
form that can be reasoned with by a computer
(Baader et al., 2003). Ontology individuals, which
are described as instances of ontology classes, repre-
sent entities described in the article and form the
nodes of a semantic graph. Ontology properties that
describe the specific relationships between those
entities form the arcs. A semantic relationship oc-
curs as a segment of a graph containing a domain
instance and a range instance linked by a property.
KDIR 2010 - International Conference on Knowledge Discovery and Information Retrieval
36
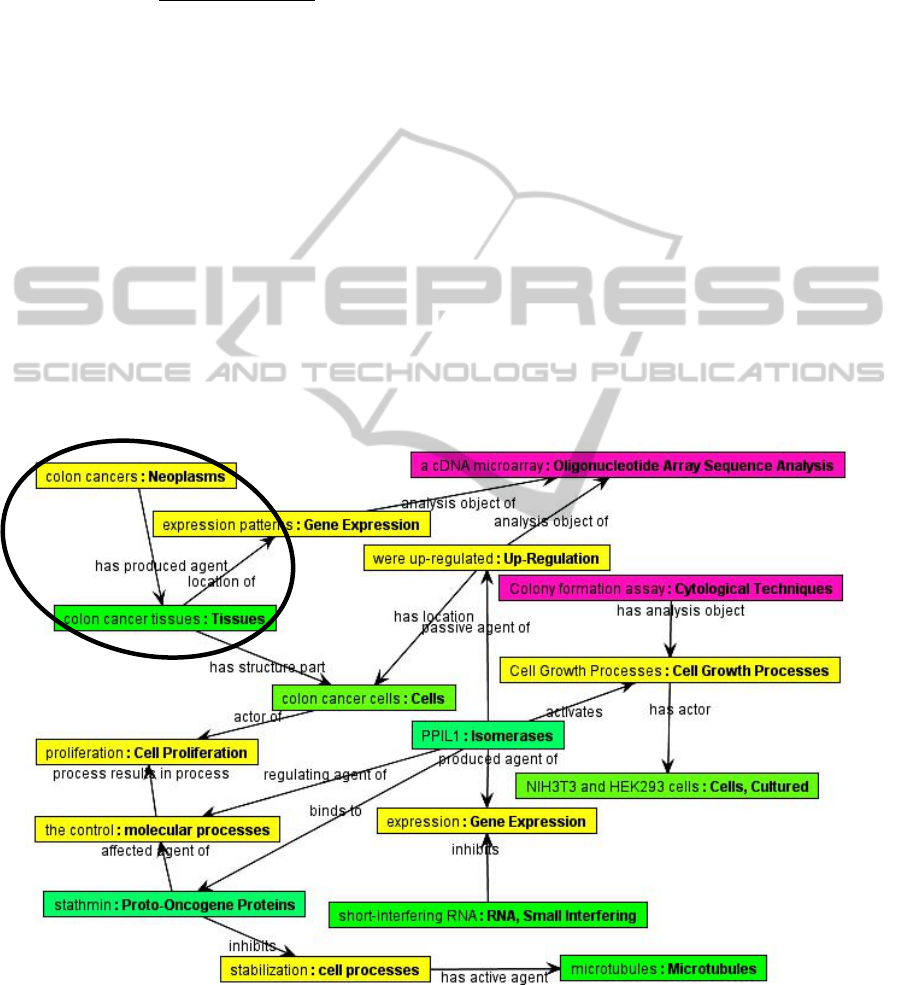
An example of a semantic graph for the article
“Over expression of peptidyl-prolyl isomerase-like 1
is associated with the growth of colon cancer cells”
(Obama et al., 2006) is shown in figure 1. The se-
mantic relationship “an instance of Neoplasms
called colon cancers has produced agent
an instance
of Tissues called colon cell tissues” (class names are
shown in bold, instance names in italics, and prop-
erty names are underlined) that forms one segment
in the semantic graph is circled.
In order to test the hypothesis that people can au-
thor computer-understandable descriptors and that
those descriptors can be used in knowledge-
intensive computing services that would otherwise
be impossible, we have created a corpus of 392 se-
mantic graphs. Each graph was created manually
based on the abstract of a research article from
MEDLINE. The 392 research articles were selected
to represent the studies of about 200 researchers in
life sciences at the University of Tokyo. The graphs
were created using the UoT ontology, which was
developed to logically structure a subset of the
Medical Subject Headings (MeSH) controlled vo-
cabulary (Kraines et al., in preparation). The subset
is made up of more than 1300 MeSH terms chosen
to cover the topics in the selected research articles
and in an introductory textbook for life sciences
used to teach undergraduates at the University of
Tokyo. The graphs have 26 classes and 34 properties
on average, so the corpus contains 13,283 individual
semantic relationships. Most of the graphs were au-
thored by undergraduate and graduate students
studying life sciences at the University of Tokyo.
Previously, we reported a technique for extract-
ing associations between specific relationships of
concepts (Guo and Kraines, 2009; Guo and Kraines,
2010a; Guo and Kraines, 2010b). A relationship
association is analogous to concept association, such
as that evidenced by term co-occurrence in article
titles, except that instead of being between singleton
concepts, the association is between semantic rela-
tionships of the form “A has specific directed rela-
tionship X with B.” Therefore, a relationship asso-
ciation is a special kind of association rule that states
“if concept A has relationship R1 with concept B,
then it is likely that concept A has relationship R2
with concept C.”
Figure 1: A slightly abridged version of the semantic graph of the article entitled “Over expression of peptidyl-prolyl isom-
erase-like 1 is associated with the growth of colon cancer cells.” Boxes show instances of classes from the ontology. The
colour of the box indicates the subsuming major upper class: yellow instances are processes, green instances are physical
entities, pink instances are investigative techniques. The text in each box gives the instance label, followed by a colon, fol-
lowed by the class name of that instance. Arrows show properties expressing the asserted relationships between instances.
The semantic relationship described in the text is circled.
GENERATING LITERATURE-BASED KNOWLEDGE DISCOVERIES IN LIFE SCIENCES USING RELATIONSHIP
ASSOCIATIONS
37

The reason for considering associations between
relationships rather than singleton concepts is as
follows. It has been observed that much of scientific
knowledge actually takes the form of specific rela-
tionships between concepts (Weikum et al., 2009).
For example, the article represented by the semantic
graph in figure 1 describes how specific isomerases
activate growth of specific cells. Therefore, a more
appropriate “unit” for scientific discovery might be a
semantic triple: a specific directed binary relation-
ship between a domain concept and a range concept.
Using semantic triples, we can extract relationship
associations such as “studies of cells that participate
in formation of cancerous tumours often focus on the
proliferation processes that those cells undergo.”
In this paper, we describe how relationship asso-
ciations can be used in a Swanson-type knowledge
discovery process. Continuing the example above, if
we find another relationship association stating that
“several studies examining the proliferation of spe-
cific cells have found that small interfering RNA
inhibits those cells,” we could combine this new
relationship association with the previous one asso-
ciating cells involved in cancerous tumour formation
with those cells participating in cell proliferation
processes to generate the hypothesis that small inter-
fering RNA might also inhibit cells involved in tu-
mour formation.
There are two major conditions for producing in-
teresting knowledge discoveries using relationship
associations. First, the classes and properties in the
ontology must be sufficiently detailed to be able to
express meaningful relationship associations. Second,
the corpus of semantic graphs must be large enough
to check that a potential discovery has not already
been reported in the literature. Unfortunately, we
only have 392 semantic graphs to work with, which
is insufficient to satisfy the second condition. The
EKOSS system is based on the idea that if the task of
authoring the semantic graphs could be distributed
over the entire scientific community, the problem of
scalability would be solved (Pico et al., 2008; Ceol
et al., 2008). However, here we have a typical
“chicken and egg” problem: in order to convince
scientists to make the effort to create the semantic
graphs, we must show their utility, but in order to
show the utility of the semantic graphs, we need a
certain minimum number of graphs to work with.
Still, we hope that our corpus of 392 semantic graphs
will be sufficient to indicate the kind of discovery
process that might be possible with a larger corpus
of graphs, thereby helping to “jump-start” a virtuous
cycle of creating and applying semantic graphs rep-
resenting research articles. We are also working to
incorporate natural language processing and machine
learning algorithms into the semantic graph author-
ing tools in order to reduce the work load and cogni-
tive overhead of the human authors.
3 GENERATING NEW
HYPOTHETICAL
RELATIONSHIP
ASSOCIATIONS
Our method for generating new relationship associa-
tions that are potential knowledge discoveries fol-
lows the basic process proposed by Swanson for the
ABC open discovery (A to B and B to C) model
(Swanson, 1990; Srinivasan, 2004). We pick up
where we left off in the previous paper with a short
list of five relationship associations that meet the
relevance criteria for “interestingness” of the asso-
ciation (Guo and Kraines, 2010a). These relation-
ship associations, shown in Table 1, form the A-B
set. We then use all of the relationship associations,
irrespective of the “interestingness” criteria, as the
B-C set, and we create all A-C relationship associa-
tions from the (A-B, B-C) pairs where the B triples
match. This gives us a set of potential knowledge
discoveries. To check that they are indeed “new”
discoveries, we match the A-C relationship associa-
tions with each of the semantic graphs in the corpus.
The A-C relationship associations that do not match
with any of the semantic graphs are potential discov-
eries that could merit further scrutiny.
Table 1: The five relationship associations we extracted
previously (Guo and Kraines, 2010a). Each triple is shown
in the form “domain class | property | range class”. The
conditional triple is separated from the consequent triple
using “>”. The connecting class is shown in bold type.
No. Relationship association
1
Flagella | has structure part |
Cytoplasmic Structures
> physical objects | interacts with |
Cytoplasmic Structures
2
Cytoplasmic Structures | has structure part | Micro-
tubules
> Chlamydomonas | has structure part |
Cytoplasmic Structures
3
Cells | passive agent of | Neoplasms
> Cell Proliferation | has active agent | Cells
4
Gene Expression | has passive agent |
Receptors, Cell Surface
> Gene Expression | has location | Neurons
5
organism parts | structure part of | Drosophila
> Growth and Development | has passive agent | or-
ganism parts
KDIR 2010 - International Conference on Knowledge Discovery and Information Retrieval
38

We divide the overall process of generating hy-
pothetical relationship associations that are potential
knowledge discoveries into three steps: 1) matching
the B triples of A-B and B-C relationship associa-
tions, 2) generating A-C relationship associations,
and 3) matching the A-C relationship associations to
the full set of semantic graphs in the corpus. We give
details for each step in the following subsections.
3.1 Matching B Triples
In the Swanson ABC model, the hypothesis is that
associations between concepts are transitive, so that
if there is an association between concept A and B
and between concept B and C, we can infer that
there may be an association between concept A and
C via the intermediary concept B. Associations are
usually predicted based on co-occurrence of the con-
cepts, e.g. in the title of a research article.
The situation with relationship associations is
slightly different. Here we have specific relation-
ships expressed between concepts, some of which
are transitive and others which are not (they may
also be reflexive or symmetric). Two relationships,
of the form (domain class has specific relationship
with range class) are linked via a shared class,
which we call the “connecting class”. As in the pre-
vious section, classes are shown in bold and proper-
ties are underlined. Thus the relationship association
is a co-occurrence of two specific relationships in-
volving a common class. Furthermore, because the
classes in the ontology are arranged in a subsump-
tion hierarchy, the actual classes of the instances of
the connecting class do not need to be the same, as
long as they are sufficiently closely related via sub-
sumption.
We also use class and property subsumption in
matching the B triples, so that for example the triple
“cell participates in
cell process” would match with
the triple “blood cell is actor of
cell proliferation”,
where blood cell is a subclass of cell, actor of
is a
subproperty of participates in
, and cell process is a
superclass of cell proliferation. Note that unlike the
original Swanson ABC model, the relationship asso-
ciations that meet the relevance criteria proposed by
(Guo and Kraines 2010a) do support directionality in
the form of “if Triple 1 occurs in a semantic graph,
then it is likely that Triple 2 will occur.” In order to
convey this directionality to the generated A-C rela-
tionship association, we also need to include the in-
verses of the relationship associations in the B-C set,
which doubles the size of the B-C set. Furthermore,
we also look at pairs where the B-C relationship as-
sociation is first and the A-B relationship association
is second, in effect matching the A and C triples.
3.2 Generating A-C
Relationship Associations
Once we find a B-C relationship association that has
a matching triple with one of the A-B relationship
associations, we use the two relationship associa-
tions to create a new A-C relationship association.
There are several ways that we can generate the new
relationship association. In the work presented here,
we connect the non-matching triples in the two rela-
tionship associations, the A and C triples, via the
connecting class in each relationship association.
This means that in addition to having a matching B
triple, the A-B and B-C relationship associations
must also have matching connecting classes.
The rationale for using this approach is as fol-
lows. A relationship association can be thought of as
an association of two typed relationships that apply
to one entity, the entity represented by the connect-
ing class. Therefore, we would interpret the A-B
relationship association “if a neoplasm process in-
volves a cell then the cell is likely to be the actor of a
cell proliferation process” as saying that cells in-
volved in neoplasm processes often are actors of cell
proliferation.
The association of relationship associations is
also interpreted through a shared class. Therefore,
the A-B relationship association shown above could
only associate with a B-C relationship association
that also has cell (or a class subsuming or subsumed
by cell) as the connecting class. For example, the
relationship association “if a bone marrow cell is
involved in
a neoplasm process, then the bone
marrow cell is likely to contain
an oncogene pro-
tein” has bone marrow cell as the connecting class,
which is a subclass of cell, so it can be associated
with the A-B relationship association. However, the
relationship association “if a bone marrow cell is
involved in
a neoplasm process, then the neoplasm
process is likely to involve
an oncogene protein”
has cell proliferation as the connecting class. Be-
cause cell proliferation is a process, which is a
branch of the ontology subsumption hierarchy that is
orthogonal to the branch containing cell, this rela-
tionship association cannot be associated with the A-
B relationship association.
Following this line of reasoning, we create new
association relationships from pairs of relationship
associations that both have a matching B triple and a
matching connecting class. Furthermore, if the actual
connecting class is different in the two relationship
GENERATING LITERATURE-BASED KNOWLEDGE DISCOVERIES IN LIFE SCIENCES USING RELATIONSHIP
ASSOCIATIONS
39
associations (as is the case in the example above),
we create two new relationship associations using
each class. Therefore, the result of the example
above with the B-C association relationship having
bone marrow cell as the connecting class would be
the two relationship associations “if a bone marrow
cell is the actor of
a cell proliferation process, then
the bone marrow cell is likely to contain
an onco-
gene protein” and “if a cell is the actor of
a cell pro-
liferation process, then the cell is likely to contain
an oncogene protein.” Of course, the second, more
general relationship association is more likely to
match with a semantic graph in the corpus and
thereby be discounted as a discovery candidate.
3.3 Matching A-C
Relationship Associations
to the Semantic Graph Corpus
We use the description logics reasoner software,
RacerPro (www.racer-systems.com), to determine
whether or not a newly generated association rela-
tionship occurs in any of the existing semantic
graphs. For each semantic graph in the corpus, we
first add that graph to the reasoner’s knowledge base
together with the ontology used to create the graph
(here the UoT ontology). Then we submit the rela-
tionship association to RacerPro as a query and ask
RacerPro to find instances in the target graph that
can bind to each of the three class variables in the
query subject to the two specified relationships. If an
independent set of binding instances can be found,
we say that the relationship association occurs in the
target graph and is therefore not a new discovery.
The process of matching relationship associations
and semantic graphs uses both logic and rule-based
inference. The logic is built into the ontology using
formalisms provided by the description logic that is
supported by the ontology specification we used
(OWL-DL). The rules are pre-defined for a particu-
lar ontology by domain experts. By using logic and
rules, we can find matches to relationship associa-
tions that are only implied at a semantic level be-
cause the reasoner can infer relationships between
instances that are implied but not explicitly stated in
the semantic graph.
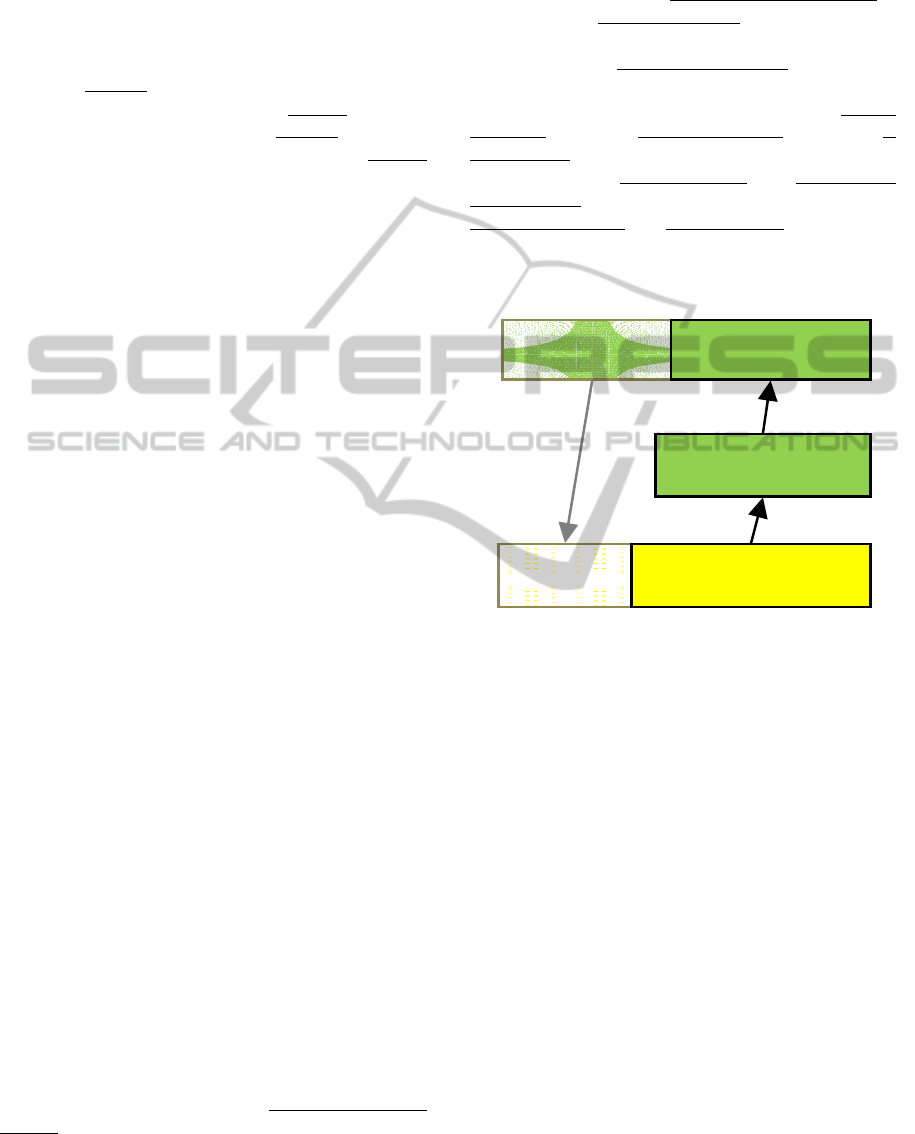
For example, consider the segment of the seman-
tic graph in figure 1 spanning two arcs between the
instance of Neoplasms called colon cancers and the
instance of Cell called colon cancer cells. The query
“find some instance of Cell that is a passive partici-
pant of some instance of Neoplasms” does not actu-
ally occur in the graph because there is no property
between colon cancer cells and colon cancers. How-
ever, as shown in figure 2, the reasoner can identify
the match between the query and the semantic graph
because the relationship is a passive participant of
is
implied by the has structure part
relationship stated
between the colon cancer cells and the colon cancer
tissues and the has produced agent
relationship
stated between the colon cancers and the colon can-
cer tissues. This match uses the rule “If A is pro-
duced by C and A has structure part B, then B is
produced by C” together with the subsumption rela-
tionship between is produced by
and is a passive
participant of and the inverse relationship between
has produced agent
and is produced by. More details
on the semantic matching process are given in
(Kraines et al., 2006; Guo and Kraines, 2008; Guo
and Kraines, 2010b).
?
Neoplasms
?
Cells
colon cancer tissues
Tissues
colon cancer cells
Cells
colon cancers
Neoplasms
has structure part
has produced agent
is passive
participant of
Figure 2: An example of semantic matching. Boxes repre-
sent instances: the first line of text gives the instance name
and the second line of text gives the instance class. Di-
rected arrows represent properties. The part outlined in
black is from the semantic graph. The part outlined in gray
is the query. Colours are the same as in Figure 1.
4 EXPERIMENTS
Using the process described above, we have con-
ducted experiments to create new relationship asso-
ciations that are potential discoveries from the rela-
tionship associations that were extracted from a set
of 392 research articles retrieved from MEDLINE
(Guo and Kraines, 2010a). In this section, we report
the results of this experiment.
4.1 Selecting the A-B Set
We hand-selected five of the 984 relationship asso-
ciations that met the relevance criteria that we speci-
fied in our previous work: the first criterion is that
the first triple must occur in no more than 40 seman-
KDIR 2010 - International Conference on Knowledge Discovery and Information Retrieval
40

tic graphs, and second criterion is that the probability
that the association query occurs when the first triple
occurs must be twice the probability that the second
triple occurs when the connecting class occurs (Guo
and Kraines, 2010a). These relationship associations,
shown in table 1, make up the A-B set of relation-
ship associations in this experiment.
4.2 Creating the B-C Set
For the B-C set, we wanted to use as many relation-
ship associations as possible, irrespective of their
“interestingness”. This is because the obvious rela-
tionship associations will be eliminated in the step
where we match the newly created A-C relationship
associations with the corpus of semantic graphs.
Therefore, we used all 4821 of the relationship asso-
ciations extracted from the corpus of semantic
graphs. Furthermore, as discussed earlier, the rela-
tionship associations can be considered to have di-
rectionality, so we also generated inverses of all of
the extracted relationship associations and added
them to the B-C set. Thus, we had a total of 9642 B-
C relationship associations to match with the five A-
B relationship associations shown in table 1.
4.3 Creating the Candidate A-C Set
The numbers of A-C relationship associations, which
are candidates for knowledge discoveries, that result
from matching the 9642 B-C relationship associa-
tions with each of the five A-B relationship associa-
tions are shown in table 2. The A-C relationship as-
sociations are generated both from pairs where the
A-B relationship association is first and from pairs
where the B-C relationship association is first. The
number of A-C relationship associations generated
for each A-B varies from 18 to 29, with an average
of 24. Therefore, on average, just 0.25 percent of the
B-C relationship associations match with each A-B
relationship association. The small number of B-C
relationship associations matching with each A-B
relationship association together with the relatively
small variance in the matches for each A-B relation-
ship association is indicative of the diversity of the
triples making up the B-C relationship associations.
4.4 Matching the A-C Relationship
Associations to Semantic Graphs
The numbers of A-C relationship associations that
were found to match with semantic graphs in the
corpus using only logic-based inference and using
both rule and logic-based inference are also shown in
table 2. By using rule-based inference in addition to
inference based on the logical properties of the
classes and properties in the DL ontology, we were
able to find matches for 1 to 3 additional A-C dis-
covery candidates. Although this is only a 10 to 20
percent increase, it indicates the value that is added
by supporting different kinds of inference in the
matching process.
On average, 53% of the A-C relationship asso-
ciations were found to already exist in the initial set
of semantic graphs, which disqualifies them as
knowledge discovery candidates. The remainder of
the A-C relationship associations are potential “dis-
coveries”. However, as we noted earlier, the number
of semantic graphs is far too small to cover all of the
semantic relationships that have been reported in the
literature. We expect that with a larger corpus of
semantic graphs, many more of the A-C candidate
relationship associations will be found to occur in
the existing literature.
Table 2: The number of A-C relationship associations that
result from matching the 9642 B-C relationship associa-
tions with the five A-B relationship associations, and the
number of those A-C relationship associations that were
found to match with semantic graphs in the corpus with
and without the application of rule-based inference.
No. Number of
A-C
relationship
associations
Number of A-
C relationship
associations
matching
without rules
Number of A-
C relationship
associations
matching with
rules
1 22 13 16
2 29 17 18
3 18 7 8
4 24 9 11
5 28 10 11
One example of an A-C relationship association
generated by the third A-B relationship association:
Cells | passive agent of | Neoplasms
> Cell Proliferation | has active agent | Cells
that did not appear in any of the graphs is:
Cells, Cultured | passive agent of | Neoplasms
> Cell Differentiation | has passive agent |
Cells, Cultured
Here we express the relationship associations with
the notation used in Table 1: “triple1 > triple2”,
where each triple is expressed as “domain class |
property | range class” and the connecting class is
shown in bold type. The B-C relationship associa-
tion is:
GENERATING LITERATURE-BASED KNOWLEDGE DISCOVERIES IN LIFE SCIENCES USING RELATIONSHIP
ASSOCIATIONS
41

Cell Proliferation | has active agent |
Cells, Cultured
> Cell Differentiation | has passive agent |
Cells, Cultured
We can interpret this relationship association to
mean that if a researcher happens to be studying
cells involved in neoplasm processes, then it might
be interesting for that researcher to look at the cell
differentiation processes of those cells.
An example resulting from the fourth A-B rela-
tionship association:
Gene Expression | has passive agent |
Receptors, Cell Surface
> Gene Expression | has location | Neurons
combined with the B-C relationship association:
Gene Expression | has location | Neurons
> Gene Expression | has passive agent |
Carboxy-Lyases
is the hypothetical relationship association:
Gene Expression | has passive agent |
Receptors, Cell Surface
> Gene Expression | has passive agent |
Carboxy-Lyases
The hypothesis generated here is that if a researcher
is studying gene expression involving cell surface
receptors, it might be interesting to look for carboxy-
lyase enzymes also involved in the gene expression.
An example resulting from the fifth A-B rela-
tionship association:
organism parts | structure part of | Drosophila
> Growth and Development | has passive agent |
organism parts
combined with the B-C relationship association:
Growth and Development | has passive agent |
Synapses
> Gene Expression | has location | Synapses
is the hypothetical relationship association:
Synapses | structure part of | Drosophila
> Gene Expression | has location | Synapses
The resulting hypothesis is that if a researcher is
studying the synapses of Drosophila, it might be
interesting to look at the gene expression located at
those synapses.
We hope that these three examples have provided
a clear demonstration of the type of scientific hy-
potheses that can be generated using the approach of
literature-based knowledge discovery from relation-
ship associations. With a larger corpus of semantic
graphs, it should be possible to extract more interest-
ing potential discoveries of new relationship associa-
tions and to check more thoroughly that those rela-
tionship associations do not already occur in the pub-
lished literature. We are currently exploring ways to
increase the size of the semantic graph corpus, e.g.
by integrating the graph authoring tools into the sci-
entific paper publication process.
5 RELATED WORK
The goal of the work presented in this paper is to
discover new knowledge or hypotheses from the
literature. Several previous research studies have
attempted to attain this goal as we mentioned in Sec-
tion 1. However, there are only a few studies that
look at knowledge discovery about specific relation-
ships between concepts.
Natarajan et al. (2006) used a combination of mi-
croarray experiments and NLP methods for extract-
ing specific gene and protein relationships, such as
inhibits and phosphorylates, from full-text research
articles, in order to discover gene interactions linked
to the protein S1P and the invasivity phenotype.
However, their sentence-based text mining results
had to be manually checked, and the problem of
gene name polysemy was noted as being particularly
difficult to resolve. They also did not appear to use
any kind of inference.
Hristovski et al. (2006) used the natural language
processing tool, BioMedLEE, to extract relationships
between genotypic and phenotypic concepts in re-
search articles, expressed in the form of “associated
with change”. They also used another NLP system,
SemRep, to extract semantic relationships in the
form of “treats”. They then used the extracted rela-
tionships to construct a “discovery pattern”, which
they defined as a “set of conditions to be satisfied for
the discovery of new relations between concepts.”
The conditions are given by combinations of rela-
tions between concepts that were automatically ex-
tracted from articles on MEDLINE. Finally, they
conducted a novelty check to find discovery patterns
that actually do not occur in the medical literature.
However, their approach suffers from the low accu-
racy of automatically extracted semantic relation-
ships and the limited number of relationship types
that could be handled.
Another technique for extracting and intercon-
necting knowledge at the relationship level is auto-
matic text summarization based on relationship ex-
traction. The CLEF (clinical e-sciences framework)
project aims to generate summaries or “chronicles”
KDIR 2010 - International Conference on Knowledge Discovery and Information Retrieval
42

of patient medical histories based on relationships
that are extracted from individual medical records
(Taweel et al., 2006). The authors indicate that infer-
ence is used in assembling individual events into
chronicles, but it is not clear if the inference is done
at the level of specific relationships between events
and entities in the records. MIAKT (Medical Imag-
ing and Advanced Knowledge Technologies) is an-
other system for automatically summarizing knowl-
edge in medical examination reports that focuses on
image annotations (Bontcheva and Wilks, 2004).
6 CONCLUSIONS
Given the tremendous rate at which the scientific
literature is increasing, new techniques are needed
for helping researchers make scientific hypotheses
that are well-based in the existing literature but have
not been reported by any previous articles. Litera-
ture-based knowledge discovery is a well-studied
approach for generating “discoveries” in the form of
potentially interesting hypotheses by finding associa-
tions between concepts that have not actually been
reported in the literature but that are implied by pre-
viously reported associations with intermediary con-
cepts. However, most existing techniques only con-
sider associations between singleton concepts.
We suggest that potentially more interesting and
meaningful hypotheses could be generated if we
considered the implied associations of specific typed
relationships between pairs of concepts or entities. In
previous work, we have developed an algorithm to
extract associations of pairs of specified relation-
ships, called relationship associations, from semantic
graphs that represent the knowledge contained in
research articles using formal “heavy-weight” on-
tologies that are based on description logics, and we
used the algorithm to extract a set of relationship
associations from a corpus of semantic graphs that
we authored for 392 articles selected from MED-
LINE.
Here, we describe an algorithm that we have de-
veloped for generating potential discoveries in the
form of relationship associations that are implied by
the extracted relationship associations but that do not
appear in any of the semantic graphs in the corpus.
We also report the results of an experiment to apply
the algorithm to the relationship associations that we
extracted previously from the 392 semantic graphs
created based on MEDLINE articles. Because each
semantic graph contains an average of 34 properties,
the corpus contains more than 13,000 semantic tri-
ples, which is comparable to the size of other major
corpora used for testing knowledge discovery appli-
cations. In fact, the number of triples that are logi-
cally entailed is easily more than 100,000. However,
even this relatively large corpus is too small to pro-
vide a good guarantee that a new relationship asso-
ciation has not actually been reported in the literature.
Still, we were able to find several new relationship
associations that at least appear to be somewhat
novel and of interest in life sciences.
The aim of this experiment using a relatively
small corpus of semantic graphs has been to provide
a demonstration of the kind of knowledge discover-
ies that could be possible if more semantic graphs
become available. In future work, we will continue
to develop the algorithm for generating knowledge
discoveries in the form of relationship associations
that are implied but not expressed in a corpus of se-
mantic graphs, and in particular we will work on
establishing additional measures of “interestingness”
for the generated relationship associations that mir-
ror the measures that we developed in our previous
work. In addition, we will continue our efforts to
realize a larger corpus of semantic graphs by devel-
oping semi-automatic methods for creating semantic
graphs and also by investigating the possibility for
integrating the semantic graph authoring approach
into the research article publication process in order
to leverage the potential for network effects in the
scientific community (Pico et al., 2008; Ceol et al.,
2008; Berners-Lee and Hendler, 2001).
ACKNOWLEDGEMENTS
The authors thank the President’s Office of the Uni-
versity of Tokyo for funding support. Hideo Ogi-
mura, Yoshihiro Okuda, and Yo Shidahara assisted
in the creation of the semantic graphs in the UoT
corpus. Takaki Makino, Yoshihiro Okuda, and Yo
Shidahara assisted in the development of the UoT
ontology.
REFERENCES
Allen, J.F., 2001. In silico veritas - Data-mining and
automated discovery: the truth is in there. EMBO Re-
ports, 2, 542-544.
Baader, F., Calvanese, D., McGuinness, D. L., Nardi,
D.,Patel-Schneider, P.F., 2003. The Description Logic
Handbook: Theory, Implementation, and Applications.
Cambridge University Press, New York.
Berners-Lee T., Hendler, J., 2001. Publishing on the Se-
mantic Web. Nature, 410, 1023-1024.
Bontcheva, K., Wilks, Y., 2004. Automatic Report Gene-
GENERATING LITERATURE-BASED KNOWLEDGE DISCOVERIES IN LIFE SCIENCES USING RELATIONSHIP
ASSOCIATIONS
43

ration from Ontologies: The MIAKT Approach. In
Proceedings of the 9th International Conference on
Applications of Natural Language to Information Sys-
tems, pp. 324-335.
Cafarella, M. J., Re, C., Suciu, D., Etzioni, O., 2007.
Structured Querying of Web Text Data: A Technical
Challenge. In Proceedings of CIDR2007.
Ceol, A., Chatr-Aryamontri, A., Licata, L., Cesareni, G.,
2008. Linking Entries in Protein Interaction Database
to Structured Text: the FEBS Letters Experiment.
FEBS letters, 582(8), 1171-1177.
Erhardt, R. A-A., Schneider, R., Blaschke, C., 2006. Status
of text-mining techniques applied to biomedical text.
Drug Discovery Today, 11(7-8), 315-325.
Gerstein, M., Seringhaus, M., Fields, S., 2007. Structured
digital abstract makes text mining easy. Nature, 447,
142.
Guo, W., Kraines, S. B., 2008. Explicit Scientific Knowl-
edge Comparison Based on Semantic Description
Matching. American Society for Information Science
and Technology 2008 Annual Meeting, Columbus,
Ohio.
Guo, W., Kraines, S. B., 2009. Discovering Relationship
Associations in Life Sciences Using Ontology and In-
ference, Proceedings of 1st International Conference
on Knowledge Discovery and Information Retrieval
2009, Madeira, Portugal, pp. 10-17, 6-8 October, 2009.
Guo, W., Kraines, S. B., 2010a. Extracting Relationship
Associations from Semantic Graphs in Life Sciences.
Communications in Computer and Information Sci-
ence (CCIS), in press.
Guo, W., Kraines, S. B., 2010b. Mining Relationship As-
sociations from Knowledge about Failures using On-
tology and Inference. 10th Industrial Conference on
Data Mining ICDM 2010, Berlin, Germany, July 12-
14, Advances in Data Mining, Lecture Notes in Artifi-
cial Intelligence (LNAI), accepted.
Hartley, J., Betts, L., 2007. The effects of spacing and
titles on judgments of the effectiveness of structured
abstracts. JASIST, 58(14), 2335-2340.
Hristovski, D., Friedman, C., Rindflesch, T. C, Peterlin, B.,
2006. Exploiting Semantic Relations for Literature-
Based Discovery. In AMIA Annu Symp Proc. 2006, pp.
349-353.
Hunter, L., Cohen, K. B., 2006. Biomedical language pro-
cessing: what's beyond PubMed? Mol Cell., 21, 589-94.
Kraines, S., 2010. An Ontology-based System for Sharing
Expert Knowledge in Life Sciences. Journal of Infor-
mation Research, in review.
Kraines, S., Guo, W., Kemper, B., Nakamura, Y., 2006.
EKOSS: A Knowledge-User Centered Approach to
Knowledge Sharing, Discovery, and Integration on the
Semantic Web. The 5th International Semantic Web
Conference, LNCS 4273, 833-846.
Kraines, S. B., Guo, W., Makino, T., Mizutani, H., Okuda,
Y., Shidahara, Y., Takagi, T., (In preparation). Trans-
forming MeSH into DL for Creating Computer-
understandable Knowledge Statements.
Langley, P., 2000. The computational support of scientific
discovery. International Journal of Human-Computer
Studies, 53, 393-410.
Natarajan, J., Berrar, D., Dubitzky, W., Hack, C., Zhang,
Y., DeSesa, C., Van Brocklyn, J. R, Bremer, E. G,
2006. Text min
ing of full-text journal articles com-
bined with gene expression analysis reveals a relation-
ship between sphingosine-1-phosphate and invasive-
ness of a glioblastoma cell line. BMC Bioinformatics,
7, 373.
Natarajan, J., Berrar, D., Hack, C. J., Dublitzky, W., 2005.
Knowledge discovery in biology and biotechnology
texts: A review of techniques, evaluation strategies,
and applications. Critical Rev in Biotech, 25, 31-52.
Obama, K., Kato, T., Hasegawa, S., Satoh, S., Nakamura,
Y., Furukawa, Y., 2006. Overexpression of peptidyl-
prolyl isomerase-like 1 is associated with the growth
of colon cancer cells. Clinical cancer research : an of-
ficial journal of the American Association for Cancer
Research, 12: 70-6.
O'donnell, M., Mellish, C., Oberlander, J., Knott, A., 2001.
ILEX: an architecture for a dynamic hypertext genera-
tion system. Nat. Lang. Eng., 7(3) 225-250.
Pico, A. R., Kelder, T., van Iersel, M. P., Hanspers, K.,
Conklin, B. R., Evelo, C., 2008. WikiPathways: Path-
way Editing for the People. PLoS Biol, 6(6), e184+.
Racunas, S. A., Shah, N. H., Albert, I., Fedoroff, N. V.,
2004. HyBrow: a prototype system for computer-aided
hypothesis evaluation. Biofinformatics, 20 (Suppl 1),
i257-i264.
Rinaldi, F., G. Schneider, K. Kaljurand, M. Hess, M. Ro-
macker, 2006. An environment for relation mining
over richly annotated corpora: the case of GENIA.
BMC Bioinformatics, 7 (Suppl 3), S3.
Seringhaus, M., Gerstein, M., 2008. Manually structured
digital abstracts: a scaffold for automatic text mining.
FEBS Lett, 582, 1170.
Smalheiser, N. R., 2002. Informatics and hypothesis-
driven research. EMBO Reports, 3, 702-702.
Srinivasan, P., 2004. Text Mining: Generating Hypotheses
From MEDLINE. JASIST, 55(5), 396-413.
Swanson, D. R., 1986. Fish oil, Raynaud’s syndrome, and
undiscovered public knowledge. Perspectives in Biol-
ogy and Medicine, 30, 7-18.
Swanson, D. R., 1990. Somatomedin C and Arginine:
Implicit connections between mutually isolated litera-
tures. Perspectives in Biology and Medicine, 33(2),
157-179.
Swanson, D. R., Smalheiser, N. R., 1997. An interactive
system for finding complementary literatures: a stimu-
lus to scientific discovery. Artificial Intelligence, 91,
183-203.
Taweel, A., Rector, A., Rogers, J., 2006. A collaborative
biomedical research system, Journal of Universal
Computer Science, 12, 80-98.
Weeber, M., Kors, J. A., Mons, B., 2005. Online tools to
support literature-based discovery in the life sciences.
Briefings in Bioinformatics, 6(3), 277-286.
Weikum, G., Kasneci, G., Ramanath, M., Suchanek, F.,
2009. Database and Information-retrieval Methods for
Knowledge Discovery. Communications of the ACM, 4,
56-64.
KDIR 2010 - International Conference on Knowledge Discovery and Information Retrieval
44
