
Repositories of Reusable Auxological (Growth)
Algorithms for eHealth
Petr Lesný
1
, Hana Krásničanová
1
, Tomáš Holeček
2
, Kryštof Slabý
1
and Jan Vejvalka
1
1
Faculty hospital in Prague Motol, V Úvalu 84, 150 06, Prague 5 – Motol, Czech Republic
2
FMS FHS Charles University, U Kříže 10, 158 00 Praha 5 Jinonice, Czech Republic
Abstract. The auxological (growth) data are suitable for being processed algo-
rithmically by specialized software. Increasing complexity of the pediatric aux-
ology requires the development of algorithm repositories, which allow the users
to select and utilize appropriate algorithms. There are many attempts to build
such repositories, however their usage is limited. In order to create the modern
repository of algorithms (not limited to the pediatric auxology), we have post-
ulated the following key principles: strong philosophical background, explicit
description of the semantic paradigm and its binding to the current scientific
paradigm, quality management, platform independency and interoperability and
trust management. Based on these principles, we have developed and validated
the indicator ontology model with strong documentation quality management,
implemented as web services under the open source license model. The refer-
ence implementation of the algorithm repository operates on the intranet of the
Faculty hospital in Prague Motol.
1 Introduction
Auxology is the science of growth. It makes no distinction between growth and de-
velopment. In this view, growth deviations do not only mean small or tall stature but
also e.g. hypotrophy, delayed puberty, alteration of closure of anterior fontanel etc.
Good knowledge of pediatric auxology (the science of growth in children) and access
to suitable reference data describing relevant population standards allow pediatricians
to make timely and correct diagnosis of growth alterations they meet in their practices
(growth diagnosis). Monitoring of the auxological parameters is also a most important
task in monitoring effect of long-term therapies in children. Auxological (growth)
data are mostly numeric and therefore highly suitable for being processed by soft-
ware.
Many specialized software tools for the analysis of growth are developed, includ-
ing standalone software tools, web calculators or sophisticated Excel spreadsheets. In
our workplace, we have developed three generations of complex tools for analyzing
the childrens growth [1-3]; the software was distributed free of charge to all of the
pediatricians in the Czech Republic to increase the quality of the diagnosis of growth
alterations in children in early stages.
Lesný P., Krásni
ˇ
canová H., Hole
ˇ
cek T., Slabý K. and Vejvalka J. (2009).
Repositories of Reusable Auxological (Growth) Algorithms for eHealth.
In Proceedings of the 1st International Workshop on Open Source in European Health Care: The Time is Ripe, pages 54-58
DOI: 10.5220/0001827900540058
Copyright
c
SciTePress
However the complexity of the growth diagnosis is continuously increasing with
new algorithms being published and validated (e.g. the weight to height analysis or
total body fat assessment). The diversity of population in the Czech Republic as new
member state of the European Union significantly increases, requiring more complex
assessment of the childrens growth (requiring e.g. the ethnic background of the child
in order to correctly evaluate the growth alterations). There are currently many at-
tempts of creating the collection of biomedical algorithms, including the auxological
ones (such as MEDAL [4], S.M.A.R.T.I.E. [5], MedCalc [6] etc.), each using differ-
ent structures and classifications for documenting of them other algorithm collections
are included in the specialized (e.g. pediatric) software, often without proper docu-
mentation. The present situation, in which the number of algorithms, reference data
and overall knowledge of the childrens growth increase requires more complex ap-
proach, such as building the algorithm repositories.
2 Materials and Methods
We have performed retrospective analysis of the current representations of algorithms
and algorithm repositories and formulated the “key principles” in the philosophy,
documentation and implementation areas, on which the successful solution should be
based. Successively, we have implemented the software and validated its usefulness
and effectiveness in the field of pediatric auxology.
Table 1. The key principles in the selection of successful solution.
Number Key principle Enables
1
Strong philosophical back-
ground
Create the ontology for description of the algo-
rithms, data, relationships between them and other
required concepts.
2
Semantic information
explicitly described and
bound to current scientific
paradigm
The inclusion of our algorithms’ collection into the
Evidence based medicine (EBM) standards.
3
Quality management
Minimize effort required to maintain the algorithm
collection and inclusion of the up-to-date know-
ledge.
4
Platform independency and
interoperability.
To minimize the time to adopt the published algo-
rithms for the end users.
5 Trust management
Support users’ decisions about procedural and se-
mantic values of individual components (e.g. the
algorithms).
55
3 Results
3.1 Philosophy
There are many existing classical (Aristotelian) biomedical ontologies and classifica-
tion systems, such as the MeSH [7], SNOMED [8] or UMLS [9], however if we want
to utilize them for description of the biomedical algorithms and their relations, we are
seriously limited. Their main problem is, that they are typically structured from the
top, whereas the concepts, utilized in the clinical applications of the algorithmic med-
icine are very ‘low-level’: none of the systems is currently able to formally represent
e.g. “the patient's body height in the morning” (the indicator commonly utilized in the
anthropometry) without substantial rearrangements and expansion of the concept tree.
We have therefore utilized the “indicator ontology”, in which the data processed
by biomedical algorithms are described (in accord with the philosophical tradition of
phenomenology represented e.g. by E. Husserl [10]) as indicators that can be trans-
formed (in a given context) into other indicators and grouped into indicator classes by
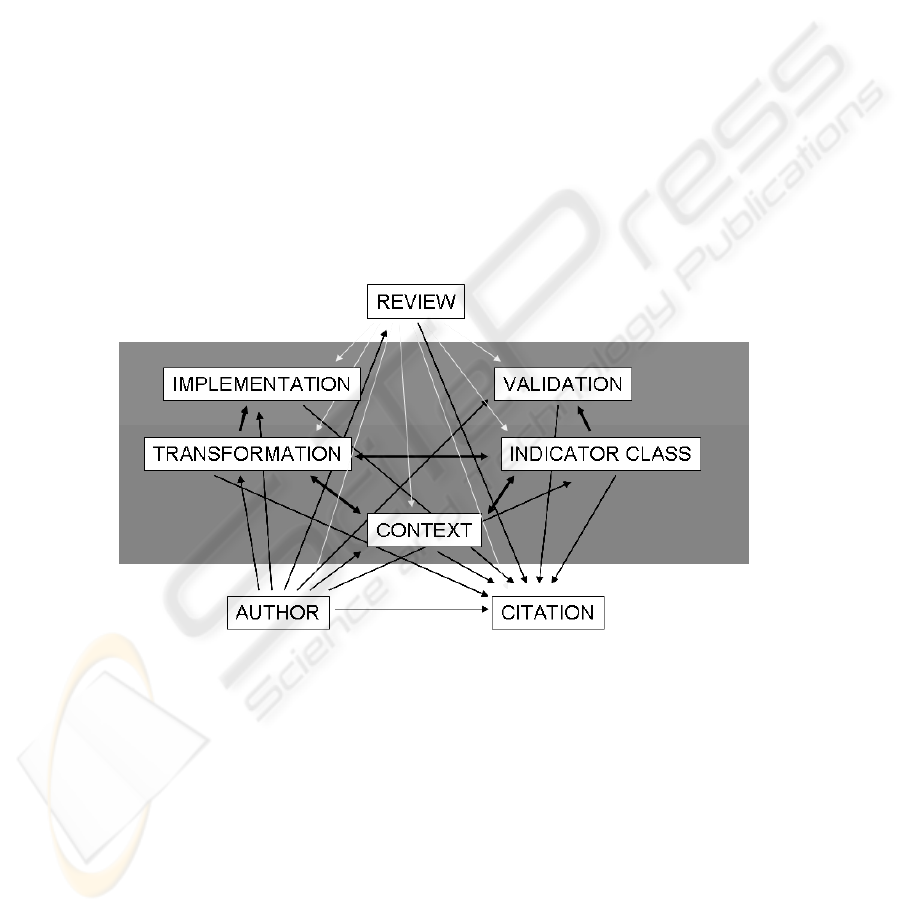
their roles in transformations. The simple representation of the indicator ontology
which shows the documentable entities (author, citation, context, transformation,
indicator class, implementation, validation and review), is on the Figure 1.
Fig. 1. The basic indicator ontology model, depicting the documentable entities in four layers
(the knowledge source layer containing the author and citation documentation, the algorithm
layer containing the transformations, indicator classes and contexts documentation, the imple-
mentation layer which contains the implementation of transformations and validation of indica-
tors and the review/trust layer)
3.2 Documentation
The biomedical algorithms should be fully documented; the semantic information
(meaning for the human user) must be explicitly described and available for
assessment and validation. The semantic information must be bound to the current
56
scientific paradigm and to evidence based medicine through predefined relations to
published and reviewed works. Optimal documentation and review paradigm is the
extension of the S.M.A.R.T.I.E. [5] model, which atomizes the semantic information
into small documentation blocks. These documentation blocks (e.g. the abstract,
method of measurement or limit conditions description) can be then organized by the
computer in order to accomplish the often conflicting requirements: (i) present the
user with full semantic information and (ii) emphasize to the user the information
relevant for the specific situation. The S.M.A.R.T.I.E. model also establishes the
basic quality management guidelines, which are similar to those utilized in the peer-
reviewed journals; they should be extended by the basic controlled documentation
management (e.g. monitoring the change of published data and ensuring the
appropriate reaction to it).
3.3 Implementation
Currently there are many possibilities of the implementation, which are summarized
in the Table 2; each of them has its advantages and disadvantages. In the biomedical
algorithm implementation (translation into the form understandable by programmers),
we require the platform independency and possibility of interoperability with other
software. We also need to review the implementation, in order e.g. to validate the
algorithms before including in specialized third party software.
Table 2. The key principles in the selection of successful solution.
Implementation type Platform
independent
Interoper-
able
Easy to use
Allows
review
Standalone software application x
Precompiled libraries x
Published code or pseudo code x x x
Web applications x x
Web services x x x
Neither of the implementation options in Table 2 alone is sufficient for the algo-
rithm repository implementation. However in order to allow user review of the algo-
rithms, the source code (or pseudo code) of the algorithms should be published, pre-
ferably together with the reference implementation. The favored reference implemen-
tation, according to Table 2, is the form of web services, which are able to easily
interoperate with other software and its usage is platform independent, together with
the published code (or pseudo code), which enables the peer reviews of the algo-
rithms’ implementation.
3.4 License
According to our key principles, the algorithms’ implementation should allow the
validation by peer review. More specifically, all the transformations of input indica-
tors to output indicators (all data processing through the algorithms) should be trans-
57

parent not only in the reference implementation, but also in all of the software prod-
ucts, which utilize the algorithms from the repository. This can be established by
adopting the open-source ‘viral’ model of software publication.
4 Conclusions
We implemented the framework for medical algorithms documentation which is
based on the key concepts presented in this work and validated its usability for docu-
menting and utilizing of the auxological algorithms [11]. The implementation is based
on the web services and interoperates with standalone Windows and Linux applica-
tions, LAMP (Linux – Apache – PHP – MySQL) web pages and even with the Mi-
crosoft Office documents (which are capable of calling the web services through the
ActiveX objects). The reference implementation is available in open source reposito-
ry
1
and also utilized in the intranet of the largest childrens’ hospital in the middle
Europe in Prague Motol.
Acknowledgements
Supported by Czech research project no. 1ET202090537.
References
1. Lesny P., Krasnicanova H.: Graphical monitoring of growth data using a computer soft-
ware Growth 1. Cesko-Slovenska Pediatrie Supplement (1996) 98.
2. Lesny P., Krasnicanova H.: Growth 2 - software for follow up of the childrens’ growth.
Prague, Maxdorf (1998).
3. Krasnicanova H., Lesny P.: Compendium of pediatric auxology 2005. Praha, Novo Nordisk
(2005).
4. Sen D.: The Medical Algorithms Project. Occupational Medicine (2005) 645.
5. Smart Medical Applications Repository of Tools For Informed Expert (S.M.A.R.T.I.E.)
and MedNotes™. IST (2000). http://www.smartie-ist.org
6. Stroud S.D., Erkel E.A., Smith C.A.: The use of personal digital assistants by nurse practi-
tioner students and faculty. J Am Acad Nurse Pract (2005) 67-75.
7. Lipscomb C.E.: Medical Subject Headings (MeSH). Bull Med Libr Assoc (2000) 265-6.
8. Cote R.A., Robboy S.: Progress in medical information management. Systematized nomen-
clature of medicine (SNOMED). Jama (1980) 756-62.
9. Lindberg C.: The Unified Medical Language System (UMLS) of the National Library of
Medicine. J Am Med Rec Assoc (1990) 40-2.
10. Husserl E. Logical Investigations, Investigation I. (translated by J. N. Finday). Routledge,
London (2001) 184.
11. Kuba M., Krajicek O., Lesny P., Vejvalka J., Holecek T.: Grid empowered sharing of
medical expertise. Stud Health Technol Inform (2006) 273-82.
1
http://medigrid.sourceforge.net
58
