
A REVIEW ON THE CURRENT SEGMENTATION
ALGORITHMS FOR MEDICAL IMAGES
Zhen Ma, João Manuel R. S. Tavares and R. M. Natal Jorge
Faculty of Engineering, University of Porto, Porto, Portugal
Keywords: Medical Image Segmentation, Algorithm Review, Pelvic Cavity.
Abstract: This paper makes a review on the current segmentation algorithms used for medical images. Algorithms are
divided into three categories according to their main ideas: the ones based on threshold, the ones based on
pattern recognition techniques and the ones based on deformable models. The main tendency of each
category with their principle ideas, application field, advantages and disadvantages are discussed. For each
considered type some typical algorithms are described. Algorithms of the third category are mainly focused
because of the intensive investigation on deformable models in the recent years. Possible applications of
these algorithms on segmenting organs and tissues contained in the pelvic cavity are also discussed through
several preliminary experiments.
1 INTRODUCTION
The developments of imaging techniques such as
Computer Tomography (CT) and Magnetic
Resonance Imaging (MRI) offer doctors with high
resolution images which have greatly assisted the
clinical diagnosis. Meanwhile medical technicians
have to process a large number of images with much
more details; Segmentation is usually a necessary
step for the task. However, manual segmentation is
very time-consuming and the results may not be
reproducible or suffer from intra-observer and inter-
observer variability. Compared with the algorithms
for common image processing, the ones used for
medical images require more concrete application
background. Priori knowledge like the imaging
procedure and the biomechanical behaviours of
organs or structures can be critical for a successful
segmentation. Also, medical images are usually
influenced by noises and partial volume effect (Zaidi,
2005), algorithms should be sophisticated enough to
handle the segmentation task. In the past few
decades, many effective algorithms have been
proposed to perform the computer-aided
segmentation. The successful implementations of
modern mathematical and physical techniques have
considerably enhanced the accuracy of the
segmentation. In the following sections, these
algorithms are classified into three categories:
algorithms based on threshold, algorithms based on
pattern recognition techniques and algorithms based
on deformable models. We focus on the main ideas
of each category and summarize their trend and
possible applications. In the end, the features of each
type are illustrated with an example of their possible
applications to segment the pelvic cavity. We also
point out that since most of the algorithms
incorporate multiple techniques, the classification of
an algorithm may not be definite.
The paper is organized as follows: In the next
section, we make a review of the algorithms; In
Section 3, we summarize the advantages and
disadvantages of each category and discuss their
possible applications to the pelvic cavity; In Section
4, we give the conclusion.
2 ALGORITHMS REVIEW
2.1 Algorithms Based on Threshold
Most of the algorithms that belong to this category
make the premise that the interested structures can
be discerned by quantifiable features, like image
intensity or gradient magnitude. Segmentation is a
procedure of searching for pixels that satisfy the
rules defined by the thresholds. Thresholds in these
algorithms can be selected manually according to
priori knowledge or automatically through image
135
Ma Z., R. S. Tavares J. and Natal Jorge R. (2009).
A REVIEW ON THE CURRENT SEGMENTATION ALGORITHMS FOR MEDICAL IMAGES.
In Proceedings of the First International Conference on Computer Imaging Theory and Applications, pages 135-140
DOI: 10.5220/0001793501350140
Copyright
c
SciTePress

information. Algorithms can be further divided to
edge-based ones, region-based ones and hybrid ones.
Thresholds in the edge-based algorithms are
related with the edge information. Structures are
depicted by edge points. Common edge detection
algorithms such as Canny edge detector (Canny,
1986) and Laplacian edge detector can be classified
to this type. Algorithms try to find edge pixels while
eliminate the noise influence. For example, Canny
edge detector uses the threshold of gradient
magnitude to find the potential edge pixels and
suppresses them through the procedures of the non-
maximal suppression and hysteresis thresholding. As
the operations of algorithms are based on pixels, the
detected edges are consisted of discrete pixels
therefore may be incomplete or discontinuous.
Hence, it is necessary to apply post-processing like
morphological operation to connect the breaks or
eliminate the holes.
The ideas of region-based algorithms come from
the observation that pixels inside a structure tend to
have similar intensities. Region growing algorithm
(Adams and Bischof, 1994) is a typical algorithm of
this type. After selecting initial seeds, algorithms
begin to search for the neighboured pixels whose
intensities are inside the intervals defined by the
thresholds and then merge them to expand the
regions. To eliminate the dependence on initial seeds
and make the algorithm automatically, statistical
information and priori knowledge can be
incorporated to the algorithms. For example, a
homogeneity criterion was introduced in (Pohle and
Toennies, 2001)
which made the region growing
algorithms adaptive for the different locations of
initial seeds and achieved success in the
segmentation of CT and MR images. However, as
the algorithms mainly rely on the image intensity
information, they are hard to handle the partial
volume effects and control the leakage.
Information used in the hybrid algorithms
combine different image cues to complete the
segmentation. Typical examples are the watershed
algorithms (Beucher and Lantuéjoul, 1979) which
combine the image intensity with the gradient
information. In the watershed algorithms, gray scale
images are considered as reliefs and the gradient
magnitude of each pixel is treated as elevation.
Watershed lines are defined to be the pixels with
local maximum of gradient magnitude. The
segmentation procedure is to construct watersheds
during the successive flooding of the gray value
relief. Due to the combination of image information,
watershed algorithms can achieve better results, but
these algorithms tend to over-segmentation
especially when the images are noisy or the objects
themselves have low signal-to-noise ratio. Hybrid
threshold-based algorithms can further combine with
other techniques to perform the segmentation (Ng, et
al, 2006).
Due to the noise influence and partial volume
effect, the edges of organs or structures in medical
images are usually not clearly defined therefore
algorithms based on threshold are seldom used alone.
2.2 Algorithms based on Pattern
Recognition Techniques
As structures in medical images can be treated as
patterns, techniques from pattern recognition fields
can be used to perform the segmentation.
Classification algorithms are the most popular ones
for the medical image segmentation. In the
following, we mainly review the supervised
classification algorithms and the unsupervised ones.
Popular techniques used by the supervised
algorithms include supervised artificial neural
network (Alirezaie, et al, 1997), support vector
machine (Wang, et al 2001) and active appearance
models (Cootes, et al, 2001). A training set is needed
to get the classifiers. Supervised artificial neural
networks (ANNs) and support vector machines
(SVMs) are non-linear statistical data modeling tools
and can be used to model complex relationships
between inputs and outputs. Weights in the classifier
are selected through optimizing energy functionals
defined by the features of structures and are updated
through processing each sample in the training set.
The extracted information from the training set
provides important cues of the structures such as
intensity, position and shape, which can be valuable
complementary information for the segmentation of
test images. Active appearance models (AAM) are
statistical models of the shape of structures. Training
samples are used to extract the mean shape, mean
appearance and define ranges of shape parameters.
Restrictions on shape parameters guarantee the
similarity between the segmentation result and the
training samples. The segmentation procedure is to
find the better positions of the shape points
according to the appearance information. Algorithms
based on classifiers have been widely applied to
segment organs in medical images like cardiac and
brain images. If properly modelled, supervised
classification algorithms can greatly enhance the
segmentation accuracy. However, supervised
classification algorithms are sensitive to the initial
conditions. To guarantee the correctness of the
results, the training set must contain enough samples
IMAGAPP 2009 - International Conference on Imaging Theory and Applications
136

and the samples should be representative and
segmented accurately.
Popular unsupervised classification algorithms
include Fuzzy C-means algorithm (Mohamed, et al,
1998), Iterative Self-organizing Data Analysis
Technique Algorithm (Wong, et al, 2002) and
unsupervised neural network (Cheng, et al, 1996).
Unsupervised classification algorithms are also
called clustering algorithms. No training set is
needed. Instead, the structure features are extracted
from the classified points. FCM algorithm comes
from C-means (CM) or K-means algorithms, where
C or K is the pre-defined number of clusters. FCM
algorithm is an iterative algorithm with the objective
to minimize the intra-cluster variation. The labelled
pixels are assigned to the nearest clusters basing on
their weighted distances to the cluster centroids, then
the cluster centroid is updated and the pixels are re-
assigned. The algorithm ends when all the pixels
have fixed labels. FCM algorithm is commonly used
for nuclear medicine and transmission image
segmentation (Mohamed, et al, 1998). Iterative Self-
organizing Data Analysis Technique Algorithm
(ISODATA) is similar to the FCM algorithm. The
main difference is ISODATA algorithm allows for
different number of clusters while the FCM assumes
that the number of clusters is known as a priori. In
medical imaging for example the positron emission
tomography (PET) scans, clustering algorithms can
be used to segment different types of tissues and
blood (Wong, et al, 2002). Unsupervised neural
networks are based on unsupervised learning, which
means the targets are the same as the inputs. The
training of weights used in classifiers is based on the
learning rule. For example the Hopfields neural
network adopts the learning rule as winner-takes-all
to simplify the selection of weights. A successful
application of Hopfields neural network to segment
the MR brain images can be seen in (Cheng, et al,
1996). Template matching algorithms and atlas-
guided algorithms which combine the prior
knowledge to asssit the segmentation are also
popular for medical image segmentation (Gindi, et al,
1993; Akselrod-Ballin, et al, 2006).
2.3 Algorithms based on Deformable
Models
Compared with the algorithms of the above two
categories, the ones based on deformable models are
more flexible and can be used for complex
segmentations. According to the representation way
of the contour, deformable models can be classified
to parametric models and geometric models. A
moving equation should be defined to drive the
initial contours to the structure boundaries.
Therefore, the procedure of these algorithms can be
viewed as a modelling of curve evolution.
The parametric deformable models have tight
relationship with the snake method (Kass, et al,
1987). Contours are sampled as discrete points and
are tracked according to their respective moving
equations. The explicit tracking has the advantage of
high computational efficiency therefore allows for
real-time applications. The moving equation for the
parametric deformable models can be derived
through either energy functional or dynamic forces.
The definition of energy functional contains two
parts: internal energy and external energy. The
internal energy aims to keep the smoothness and
regularity of the contour and is usually defined
through the geometric properties of the contour such
as length, area and curvature; the external energy
aims to drive the contour to the right position and
the definition is based on image information. Using
calculus of variations, we can derive an Euler-
Lagrange equation of the energy functional which
states that the balancing equilibrium of the moving
contour under the external forces and the internal
forces is the position of structure boundary. Through
adding the time variable to the Euler-Lagrange
equation, we then get the dynamic moving equation.
Priori knowledge can be easily incorporated to the
procedures of parametric models. Snake method is
the first deformable model applied to the medical
image segmentation. The traditional snake method
relies on the gradient information therefore is
sensitive to the initial position of the contour. The
contour must be placed to the positions near to the
structure boundary so that the external forces are
strong enough to attract the contour. Otherwise the
contours may shrink or stop at wrong positions. The
later proposed algorithms (Cohen, 1991; McInerney
and Terzopoulos, 1995; Xu and Prince, 1998) tried
to eliminate the dependence on the initial conditions
and noise influence. Cohen (Cohen, 1991) added a
balloon force to the external forces to make the
contour inflation or deflation even if the gradient
field is weak. Xu and Prince (Xu and Prince, 1998)
replaced the gradient field with the gradient vector
field (GVF) which achieves the same capture effect
near the structure boundary while changes slowly in
other regions.
The geometric deformable models are based on
the level set method (Osher and Sethian, 1988)
which was initially proposed to handle the
topological changes during the curve evolution. The
main idea of the level set method is to implicitly
A REVIEW ON THE CURRENT SEGMENTATION ALGORITHMS FOR MEDICAL IMAGES
137

embed the moving contour into a higher dimensional
level set function and view the contour as its zero
level set. Then, instead of tracking the contour
points, we can track the zero level set of the level set
function. The advantage of doing so is that
topological changes can be naturally handled and the
geometric properties of the contour such as normal
vector and curvature can be calculated implicitly.
Therefore, the computational complexity is
decreased. Like the parametric deformable models,
speed functions should be properly defined. Malladi,
et al. (Malladi, et al, 1993) and Caselles, et al.
(Caselles, et al, 1997) first applied the level set
methods to medical images. Malladi’s model used
the gradient information as a stop criterion. The
definition of the speed is intuitive: when the contour
moves to the structure boundary, the increase of
gradient magnitude decreases the speed value
therefore slows down the contour. However, this
speed model suffered from leakage due to its mere
dependence on the gradient magnitude.
Unlike Malladi’s model, geodesic active contour
algorithm (Caselles, et. al, 1997) treated the
segmentation as an optimization problem of finding
the minimal distance curve. The moving equation of
GAC is also derived through energy functional.
While instead of solving directly the moving
equation, the contour is embedded in a level set
function and the moving equation then becomes a
level set equation. Geodesic active contour
algorithm showed a tight relationship between the
parametric model and the geometric model. The
introduction of level set representation in GAC
makes the algorithm flexible to handle the
topological changes. GAC and the later improved
GAC algorithms are applied to process the MR, CT
and ultrasound images like the tumour detection and
cardiac segmentation (Paragios, 2002). Another
popular geometric model Chan-Vese’s model is
based on a simplified version of Mumford-Shah
energy model (Chan and Vese, 1999). The most
appreciable advantage of Chan-Vese’s algorithm is
that it can obtain a boundary of discrete points,
which is quite useful for medical image applications
when the interested structures are represented by
discrete pixel clusters and have no clear definition of
boundaries.
3 DISCUSSIONS
When the interested structures have distinctive
quantifiable features, using threshold-based
algorithms is effective. Threshold-based algorithms
do not need complex operations therefore are
computationally efficient. However, due to their
dependence on thresholds, these algorithms are
sensitive to noises, hard to combine with spatial
information and difficult to be applied to multi-
channel images. As medical images are usually
noisy and suffer from intensity inhomogeneity, the
segmentation results of threshold-based algorithms
are usually far from satisfaction. Therefore these
algorithms are seldom used alone. Instead they are
often used as an efficient pre-segmentation step.
Compared with threshold-based algorithms, the
ones based on pattern recognition techniques can
better utilize structure information therefore can
achieve satisfied results. When the structures in
medical images are regular and not much influenced
by noises, applying pattern recognition techniques is
effective. However, like the threshold-based models,
pattern recognition models are also sensitive to
noises. The results of these algorithms may depend
on their initialization step. For the classifier-based
algorithms, the segmentation results depend on the
size of the training samples and the correctness of
the manual segmentations. For the clustering
algorithms, the number of clusters, the position of
the initial points and the parameters should be
properly selected. The lack of incorporating spatial
characteristics is also an obstacle. Due to the large
shape variations in medical images, the applications
of these algorithms are constrained.
Due to the advantages of being able to handle
structures with complex topology, easy to
incorporate with other techniques, sub-pixel
accuracy, noise insensitive and intuitive interaction
mechanisms, the deformable models are intensively
investigated in the last few decades. Parametric
deformable models have high computational
efficiency and are easy to incorporate with other
techniques; Geometric deformable models have the
advantage of naturally handling the topological
changes. For the medical image segmentation, using
parametric model or geometric model depends on
the applications. In general, when structures have
large shape variety or complicated topology,
geometric deformable models are preferred; when
the interested structures have open boundaries or the
structures are thin or the algorithms need real-time
operations, parametric models are preferred.
However, deformable models usually contain certain
number of parameters. To select proper parameters
is critical to the final segmentation results while this
is usually a time-consuming job.
CT images and MR images are widely used
modalities to study the organs in the pelvic cavity.
IMAGAPP 2009 - International Conference on Imaging Theory and Applications
138
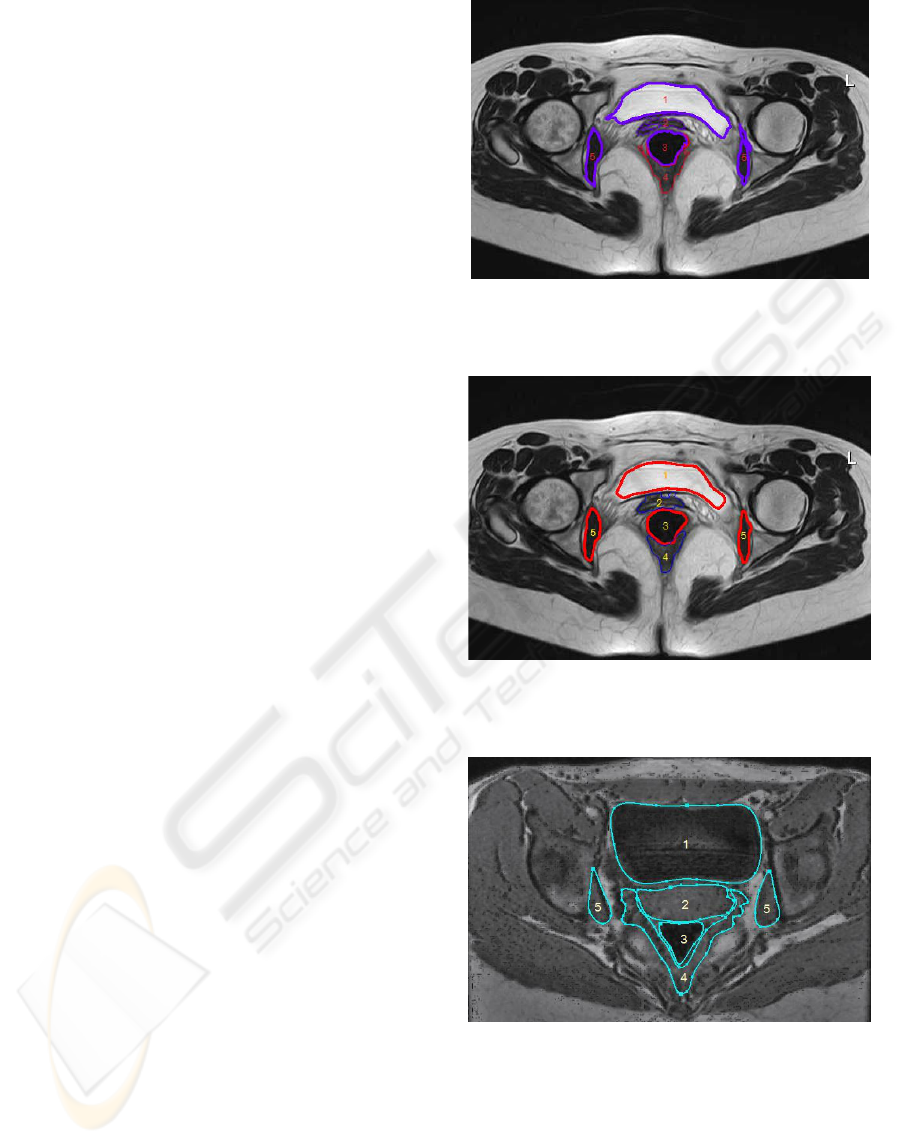
Figure 1 and Figure 3 illustrate the two imaging
models. Segmentation task includes sketching
bladder, urethra, vagina, rectum and pelvic floor.
The complex anatomical structures and the inter-
connectivity between organs make the segmentation
difficult to perform. We do not intend to propose
new algorithms here but use this example to make a
preliminary discussion of possible applications of
the three categories of algorithms.
Figure 1 illustrates the segmentation result of
region grow algorithm; Due to the high resolution
and signal-to-noise ratio, bladder and vagina can be
segmented successfully. While the boundary of the
right obturator internus is leaked due to the influence
of partial volume effect. Figure 2 illustrates the
segmentation result of geodesic active contour
algorithm. The boundaries of the obturator internus
are correctly segmented and the boundary of vagina
is not leaked in the upward direction. This is due to
the restriction of smoothness of the moving contours.
The structure boundaries are more regular than the
ones in Figure 1. While also due to this restriction,
small details of structure boundaries are eliminated
so that the boundaries of bladder and levator ani are
not complete. For the two segmentations, both
boundaries of levator ani are not satisfied. An
example of manual segmentation of levator ani is
illustrated in Figure 3. In fact, the segmentation of
levator ani muscles needs prior shape information
therefore a correct segmentation depends on
information beyond the presented images. This
feature is the advantage of the algorithms based on
pattern recognition techniques. For the CT images
which contain large amount of noises and low
spatial resolution of soft tissues, the priori shape
information is more important. The result of
applying watershed algorithm to images in Figure 3
is illustrated in Figure 4, from which we can see the
noise influence, the discontinuity and the difficulties
in this application. A possible way to design a new
algorithm can be incorporating the comparative
distances of different organs and use the prior shape
information to constrain the result.
4 CONCLUSIONS
As pointed out in Section 1, most of the algorithms
combine multiple segmentation techniques and use
diverse image cues to improve the segmentation
results. Therefore, a definite classification of an
algorithm may be infeasible. In this paper, we
classify the current algorithms into three categories
and summarize their features. From the discussions
we can see that each category has its suitable
Figure 1: Segmentation results of MR image using region
growing algorithm: 1-bladder, 2-vagina, 3-rectum, 4-
levator ani, 5-obturator internus.
Figure 2: Segmentation results of MR image using
geodesic active contour algorithm: 1-bladder, 2-vagina, 3-
rectum, 4-levator ani, 5-obturator internus.
Figure 3: Manual segmentation results of CT image: 1-
bladder, 2-vagina, 3-rectum, 4-levator ani, 5-obturator
internus.
application fields. For a concrete medical image
segmentation task, researchers should combine the
application background and practical requirements
to design proper algorithms. Accuracy, complexity,
efficiency and interactivity of a segmentation
algorithm should all be the considered factors.
A REVIEW ON THE CURRENT SEGMENTATION ALGORITHMS FOR MEDICAL IMAGES
139

Figure 4: Segmentation results of CT image using
watershed algorithm.
ACKNOWLEDGEMENTS
This work was partially done in the scope of the
project “BIOPELVIC-Study of Female Pelvic Floor
Disorders”, with reference PTDC/SAU-
BEB/71459/2006, financially supported by
Fundação para a Ciência e a Tecnologia of Portugal.
REFERENCES
Adams, R., Bischof L., 1994. Seeded Region Growing.
IEEE Trans. on Pattern Anal Mach. Intelligence,
16:641-47.
Akselrod-Ballin, A, Galun, M, et al, 2006. Atlas Guided
Identification of Brain Structures by Combining 3D
Segmentation and SVM Classification. Int. Conf Med
Image Comput Comput Assist Interv., 209-16.
Alirezaie, J., Jernigan, M. E., Nahmias, C., 1997. Neural
Network-Based Segmentation of Magnetic Resonance
Images of the Brain, IEEE Trans. on Nuclear Science,
44:194-8.
Bezdek, J. C., Hall, L. O., Clarke, L. P., 1993. Review of
MR Image Segmentation Techniques Using Pattern
Recognition, Med. Phys., 20:1033-48.
Canny, J., 1986. A Computational Approach to Edge
Detection. IEEE Trans. Pattern Analysis and Machine
Intelligence, 8:679-714.
Caselles, V., Kimmel, R., Sapiro, G., 1997. Geodesic
Active Contours. Int. J. of Comp. Vision, 22:61-79.
Chan, T., Vese, L. A., 1999. An Active Contour Without
Edge. Int. Conf. Scale-Space Theories in Computer
Vision, 141-151.
Cheng, K. S., Lin, J. S., Mao, C. W., 1996. The
Application of Competitive Hopfield Neural Network
to Medical Image Segmentation. IEEE Trans. on Med.
Img., 15:560-7.
Cohen, L. D., 1991. On Active Contour Models and
Balloons. CVGIP: Image Understanding, 53:211-8.
Cootes, T. F., Edwards, G. J., Taylor, C. J., 2001. Active
Appearance Models. IEEE Trans. Pattern Analysis
and Machine Intelligence, 23: 681-85.
Davis, L. S., 1975. A Survey of Edge Detection
Techniques. Comp. Graphics and Image Processing,
4:248-70.
Flitti, F., Collet, C., Joannic-Chardin, A., 2005.
Unsupervised Multiband Image Segmentation Using
Hidden Markov Quadtree and Copulas. IEEE Int. Conf.
on Image Processing, 2:634-37.
Gindi, G., Rangarajan, A., Zubal, G., 1993. Atlas-Guided
Segmentation of Brain Images via Optimizing Neural
Networks. Proc. SPIE Biomedical Image Processing
IV, 1905-58.
Kass, M., Witkin, A., Terzopoulos, D., 1987. Snakes:
Active contour models. Int. J. of Comp. Vision, 1:321-
31.
Malladi, R., Sethian, J. A., Vemuri, B., 1993. A Topology
Independent Shape Modeling Scheme. SPIE Conf. on
Geometric Methods in Computer Vision II, 2031:246-
58.
McInerney, T., Terzopoulos, D., 1995. A Dynamic Finite
Element Surface Model for Segmentation and
Tracking in Multidimensional Medical Images with
Application to Cardiac 4D Image Analysis.
Computerized Medical Imaging and Graphics, 19:69-
83.
Mohamed, N. A., Ahmed, M. N., Farag A., 1998.
Modified Fuzzy C-mean in Medical Image
Segmentation. Proceedings of the 20th Annual
International Conference of the IEEE
, 3:1377-80.
Ng, H. P., Ong, S. H., et al, 2006. Medical Image
Segmentation Using K-Means Clustering and
Improved Watershed Algorithm. IEEE Southeast
Symposium on Image Analysis and Interpretation, 61-
5.
Osher, S., Sethian, J., 1988. Fronts Propagating with
curvature-dependent speed: algorithms based on
Hamilton-Jacobi formulations. J. Comp. Phys., 79: 12-
49.
Paragios, N., 2002. A Variational Approach for the
Segmentation of the Left Ventricle in Cardiac Image
Analysis, International Journal of Computer Vision,
50:345-62.
Pohle, R., Toennies, K. D., 2001. Segmentation of
Medical Images Using Adaptive Region Growing,
SPIE, 1337-46.
Wang, S., Zhu, W. Y., Liang, Z. P., 2001. Shape
Deformation: SVM Regression and Application to
Medical Image Segmentation, Eighth International
Conference on Computer Vision, 2:209-16.
Wong, K. P., Feng, D., et al, 2002. Segmentation of
Dynamic PET Images Using Cluster Analysis. IEEE
Trans. on Nuclear Science, 40:200-07.
Xu, C. Y., Prince, J. L., 1998. Snakes, Shapes, and
Gradient Vector Flow. IEEE Trans. on Image
Processing, 7:359-69.
Zaidi, H., 2005. Quantitative Analysis in Nuclear
Medicine Imaging, Springer.
IMAGAPP 2009 - International Conference on Imaging Theory and Applications
140
