
An Image Mining Medical Warehouse
Sara Colantonio
1
, Igor B. Gurevich
2
, Ovidio Salvetti
1
and Yulia Trusova
2
1
Institute of Information Science and Technologies (ISTI)
Italian National Research Council (CNR), Via Moruzzi 1, 56124, Pisa, Italy
2
Dorodnicyn Computing Center of the Russian Academy of Sciences
Vavilov str. 40, 119333 Moscow, Russian Federation
Abstract. Advances in medical imaging technologies have assured the availabil-
ity of more and more precise and detailed images whose analysis has became a
necessary step in the diagnostic, prognostic and monitoring processes of main
pathologies. Such development has stressed the need for advanced systems that
are not limited to storage and management but include intelligent representa-
tion and retrieval of images. In this paper, we report current results of a medical
warehouse we are developing for mining medical images, thus offering medical
experts and researchers the possibility of storing, retrieving, analyzing and inves-
tigating biomedical images to discover novel knowledge relevant to diagnostic
processes.
1 Introduction
Nowadays, the field of visual information technology is one of the main sources for
knowledge representation and understanding. This is dramatically true within the med-
ical sciences where the importance of image content makes more urgent the call for
proper and efficient storage, management and usage of images.
Specialized systems, termed Picture Archivingand Communication Systems-PACS,
have been introduced within the Health Information Systems for the acquisition, stor-
age, transmission, processing and display of digital, multi-modal medical images [8].
PACS provide efficient and cost-effective means for off-site retrieving, examining and
reporting diagnostic images, thus allowing for tele-diagnosis, second opinion consul-
tancy and distance education. However, these large collections of images hide rich
information highly useful in the diagnostic and monitoring processes, which requires
advanced and intelligent techniques to be extracted. Image Mining deals with the ex-
traction of implicit, semantically meaningful knowledge, image data relationships, or
other patterns which are not explicitly stored in the image databases, and which con-
nect images to non imagery contextual data [15]. Research in image mining is still in
its early stages, although it relies on rather assessed disciplines such as computer vi-
sion, image processing, image retrieval, data mining, machine learning, database, and
artificial intelligence. The fundamental challenge in image mining is to determine how
low-level representation contained in a raw image or image sequence can be processed
to identify high-level, novel information and relationships among data.
Colantonio S., B. Gurevich I., Salvetti O. and Trusova Y. (2008).
An Image Mining Medical Warehouse.
In Image Mining Theory and Applications, pages 83-92
DOI: 10.5220/0002341400830092
Copyright
c
SciTePress

Two main typologies of image mining frameworks can be individuated: function-
driven, which are focused on the functionalities of the components to be integrated, and
information-driven, which are designed as a hierarchical structure with special empha-
sis on the information needs at various levels of the hierarchy.
Within the medical field, some image mining systems have been presented [13], es-
pecially within histology and cytology [3], [4], [10]; however the main focus of their
development has been reserved to the problem of image retrieval by content, thus miss-
ing the real interesting challenge of extracting new knowledge. Exploiting experiences
matured within the EU Network of Excellence MUSCLE (Multimedia Understand-
ing through Semantics, Understanding and Learning), the STREP EU project HEART-
FAID (A knowledge based platform of services for supportingmedical-clinical manage-
ment of the heart failure within the elderly population) and the Italian-Russian bilateral
project, we are working on the development of an Image Mining Medical Warehouse
(IMMW) which is meant to supply all the image mining functionalities, ranging from
image storage to novel knowledge discovery. IMMW has been designed by integrating
and extending the infrastructure we have developed within the MUSCLE initiative for
managing multimedia metadata (the so-called 4M infrastructure) [1] by integrating a
semantic apparatus for suggesting suitable image analysis algorithms and appropriate
decisions on medical diagnosis.
In this paper, current results of the design activity are presented since interesting
aspects have come forth as worthy of discussion. The main components of IMMW are
also introduced and future activities discussed.
2 The Infrastructure for an Image Mining Medical Warehouse
IMMW infrastructure has been conceived for aiding the investigation and diagnostic
processes of medical problems, by providing local and remote access to known cases,
the facility for retrievingimages by content, and the possibility of mining image patterns
relevant to medical decision making, e.g., diagnoses and prognoses.
Several practical scenarios can be supplied for highlighting the usefulness of IMMW
functionalities:
· Case-based reasoning: a physician is presented with magnetic resonance imaging
(MRI) data of a patient’s brain, but imaged patterns raise some uncertainty in the
diagnosis. Retrieving similar cases according to patient’s information and the im-
agery data can help him making a final decision by investigating other physicians’
diagnoses;
· Exploration of concurrentsituations: a histopathologist researcher is experimenting
with a new marker on breast tissue. For comparing its performance, he can search
for the effects of the same marker on different tissues, e.g., on liver tissue, or for
other markers resulting in the same effects on breast tissue;
· Extraction of new knowledge: a cardiologist wants to explore the prognostic value
of some parameters extracted from imaging data, e.g., of the ejection fraction com-
puted from cardiac MRI. Applying data mining techniques on opportunely defined
patterns, he can find out the relationship between prognosis and the set of parame-
ters.
8484
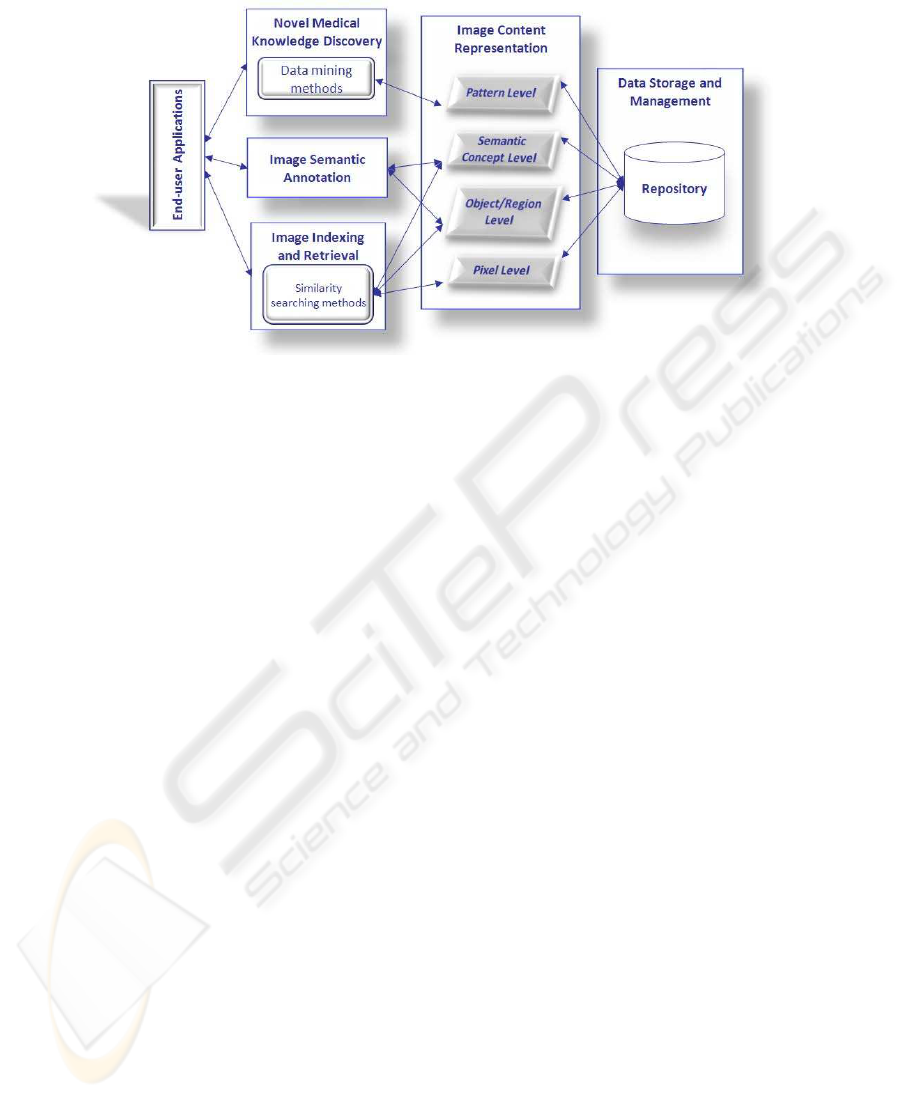
Fig.1. Main functionalities of the Image Mining Medical Warehouse.
IMMW has been functionally designed as shown in Figure 1, also considering a
hierarchy of information levels. Actually, along with the data storage and management
inside a repository, we identified three main and interrelated end-users functionalities:
· Image indexing and retrieval, for finding images ranked in accordance to some re-
quirements on their content. Retrieval can be performed by means of explicit text
query or by supplying a reference image. Similarity measures are applied to appro-
priate image representations for identifying the relevant images to be retrieved;
· Image semantic annotation, for defining a list of semantic keywords to be associated
to image and used for their retrieval, in particular for text queries. A structured
terminology is supplied for aiding annotations by the users;
· Novel medical knowledge discovery, for extracting valid, novel and understandable
knowledge about the diagnostic, prognostic and monitoring processes. Advanced
data mining methods can be applied to patterns built by correlating features ex-
tracted from images and domain concepts.
These functionalities rely on the fundamental process of representing image infor-
mative content by extracting a set of meaningful features and interpreting them. Within
IMMW, four levels of representation can be conceptually identified, as suggested in
[15]:
i. The Pixel level consists of the raw image information such as image pixels and
global primitive image features computed on them;
ii. The Object level deals with object or region information extracted on the basis of
the primitive features;
iii. The Object level deals with object or region information extracted on the basis of
the primitive features;
8585

iv. The Pattern level incorporates domain related data and the semantic concepts ob-
tained from the image data to discover underlying domain patterns and knowledge.
The first level yields a coarse representation model which is built with low-level
features computed on pixel data, such as color and texture, and represents the basic
information for retrieving images by content. Being global, such model lacks impor-
tant concepts of object or region, thus is useless for answering queries such as “find
brain MRI that highlights an intracranial aneurysm”. The second level is conceived for
filling this lack: it often requires an image segmentation process to assure a better ac-
curacy of specific, sometimes domain-dependent features computed for representing
the objects/regions contained in the images. A further elaboration involves an object
recognition process which can be also seen as part of the third level: through pattern
recognition and image categorization/understanding methods, semantic concepts be-
longing to the medical domain knowledge are associated to images. The fourth level
is of key importance for image mining purposes and is concerned with the integration
of high level information extracted from imaging and contextual data of the medical
domain in order to mine novel patterns relevant for the medical decision problems.
IMMW conceptual design is being developed by extending the 4M infrastructure
which was designed aiming at defining, maintaining and exchanging multimedia meta-
data and data sets. 4M was developed by using (i) standardized Semantic Web tech-
nologies promoted by the W3C office, since they can facilitate the overall vision of
distributed, machine readable metadata; (ii) multimedia metadata standards, in particu-
lar the MPEG-7 standard; and (iii) open-source software.
The 4M is being extended by adding a semantic apparatus, constituted by a suite of
ontologies which assure several advantages as means for:
· structuring a universe of discourse by formalizing domain knowledge;
· improving and increasing the amount of domain knowledge through reasoning and
inferring new knowledge;
· combining image understanding methods for the extraction of domain semantic
concepts;
· semantic retrieval and annotation of images by using domain concepts.
The main components of IMMW, as sketched in Figure 2, are:
· a repository for storing, accessing and retrieving images and information extracted
at different levels from them;
· a suite of ontologies including specific domain ontologies related to medical prob-
lems and a general image understanding ontology;
· a collection of image analysis algorithms for image processing, analysis, recogni-
tion and mining;
· a user interface for accessing, uploading, browsing and annotating images.
Parts of the last three components have been already developed inside the 4M (the
one highlighted in Figure 2). Currently the extension into the IMMW is being conducted
working in parallel on the different components, as described in the following sections.
The integration is performed at the level of the user interface.
8686

Fig.2. Main components of IMMW. The coloured ones have been developed within the 4M.
2.1 The Suite of Ontologies and Image Processing Algorithms
Over the last years, ontologies have demonstrated to be an important and useful instru-
ment for representing, sharing and reusing knowledge. Evidences for that are: ontology
languages allow for expressing rich semantics and provide reasoning capabilities, e.g.,
the Ontology Web Language (OWL) [12] family of languages is based on Descrip-
tion Logic, thus, besides allowing for reasoning capabilities, it assures a well-founded
knowledge base; languages and tools can be easily found for free and based on standard
Semantic Web technologies (OWL, RDF, etc.). This can provide user friendly capabil-
ity to build ontologies and also makes it easier to share the results and re-use knowledge
provided by others.
Within the IMMW, the suite of ontologies contains:
· Domain ontologies, defined for representing domain knowledge;
· Image Understanding ontologies, devoted to the semantic formalization of image
processing, representation, classification and recognition techniques.
Domain ontologies have been inserted for supporting the concept level represen-
tation of image content and for aiding the users’ annotation process. So far, we have
developed an ontology of the cytopathology domain [6], while two ontologies regard-
ing the cardiologic and the breast oncology domains are under development [5].
The image understanding ontologies are being developedbasing on the Image Anal-
ysis Thesaurus [2], which has been defined at the Scientific Council “Cybernetics” of
the Russian Academy of Sciences and detailed later at the Dorodnicyn Computing Cen-
ter of the Russian Academy of Sciences [7]. Currently, the available ontologies contain
8787

Fig.3. An excerpt of image processing and features ontologies.
concepts related to image general information, and techniques for image processing and
features extraction [6]. The MPEG-7 ontology [11], already used in the 4M, has been
included and re-arranged within the feature ontology. An excerpt of the classes is shown
in Figure 3.
The ontological models have been equipped with a library of algorithms that al-
lows for the application of image transformation methods and for the extraction of the
features.
2.2 The Infrastructure for Image Management
For adequately handling the medical images, a management infrastructure has been
set up exploiting the existing components of the 4M, i.e. the repository and the user
interface.
The repository has been developed as an XML database, in order to assure the high-
est image representation flexibility. Actually, the internal representation of features can
be directly inserted into the database and data structures should be extended only to
include additional extracted features. For the implementation, an open source, native
XML database, eXist [9], has been selected, since it provides efficient, index-based pro-
cessing, automatic indexing, extensions for full-text search, and a Java interface. Java
classes have been implemented for querying the collections using XQuery language
[14] in order to extract low-level features and select image objects by similarity.
An interface has been also implemented to search for images in the database. Given
that an URI (Unique Resource Identifier) is a basic building block for Semantic Web
applications, we denote every multimedia object by a unique identifier, named Medi-
aURI, that includes the type of the object and a hash of the object content. Through a
MediaURI, any multimedia object is univocally identified and can be accessed in our
XML database.
8888

Fig.4. Two screen shots of the warehouse infrastructure showing excerpts of the mammographic
(upper) and cytological (lower) images collections.
3 Discussion and Future Activities
Current results of the IMMW consist in an infrastructure which allows for: (i) image
upload; (ii) image analysis to produce a features representation; (iii) image storage with
MPEG-7 metadata in the XML database; (iv) image retrieval by content.
The medical domains currently considered are cytopathology, histology, cardiology
and oncology. In Figure 4 small extracts of two imaging collections stored into the
IMMW are shown, i.e. the mammography and cytology.
The content-based retrieval is performed by similarity on extracted features. Figure
5 shows the results of two queries by reference image. Images are ranked according to
the similarity value.
Image annotation is a core functionality supplied by the infrastructure; in particular
annotations can be added to image regions. The annotation facility allows the user to
select a region and insert textual annotation which is stored in RDF format. Future
development will guide the annotation by supplying domain terminology (i.e. concepts
contained in the domain ontologies). Figure 6 shows the annotation of a cytological
image and the resulting RDF code. Annotations are summarized and reported onto the
annotated regions (see Figure 7).
8989

Fig.5. Results of the content based retrieval: the upper left image is the query image and the
upper right is the more relevant.
Future activities will consist in the completion of the ontology suite and its full in-
tegration with the library of algorithms for image understanding at high semantic level.
Furthermore, the actual data mining facility will be inserted allowing for the discovery
of novel medical knowledge. An ambitious goal will be the integration of the IMMW
with PACS for supporting medical researchers in their routine activity workflows.
Acknowledgements
The authors would like to thank Marco Tampucci for his valuable work for develop-
ing, extending and maintaining the 4M infrastructure. The work was partly supported
by the cooperative grant within agreement between Italian National Research Council
and Russian Academy of Sciences, by EU Network of Excellence MUSCLE - FP6-
507752, by the Russian Foundation for Basic Research (projects nos. 06-01-81009,
06-07-89203), by the EU STREP project HEARTFAID (FP6-IST-2004-027107,07-07-
13545), and by the project no. 2.14 of the Program of the Presidium of the Russian
Academy of Sciences “Fundamental Problems of Computer Science and Information
Technologies”.
9090
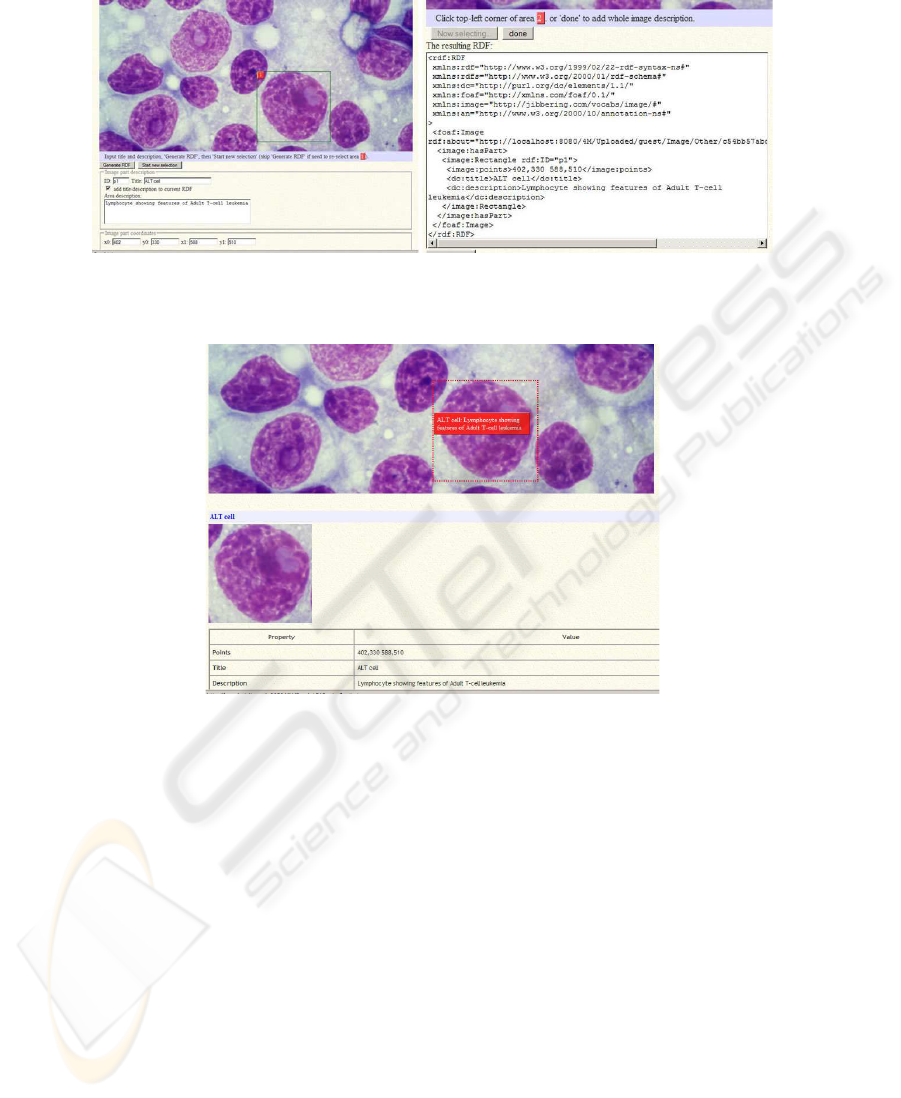
Fig.6. The annotation facility: (left) textual descriptions can be associated to image regions,
(right) the correspondign RDF code is automatically created.
Fig.7. Summary of the annotation of Figure 6.
References
1. Asirelli, P. Little, S., Martinelli M., Salvetti, O.: MultiMedia Metadata Management: a Pro-
posal for an Infrastructure. In SWAP 2006, Semantic Web Technologies and Applications,
Dec.18-20, Pisa, Italy (2006)
2. Beloozerov, V.N., Gurevich, I.B., Gurevich, N.G., Murashov, D.M., Trusova, Y.O.: The-
saurus for Image Analysis: Basic Version. Pattern Recognition and Image Analysis, Vol.
13:4 (2003) 556-569
3. Cebron, N .and Berthold, M.R.: Mining of Cell Assay Images Using Active Semi-Supervised
Clustering. In ICDM 2005 Workshop on Computational Intelligence in Data Mining (2005)
63-69
4. Chen, W., Meer, P., Georgescu, B., He, W., Goodell, L.A., Foran, D.J.: Image Mining for
Investigative Pathology Using Optimized Feature Extraction and Data Fusion. Computer
Methods and Programs in Biomedicine, Vol. 79 (2005) 59-72
9191

5. Chiarugi, F., Colantonio, S., Conforti D., Martinelli, M., Moroni, D., et al.: Decision Support
and Signal & Image Mining in Heart Failure. In HEALTINF07, Madeira, Portugal (2007)
6. Colantonio, S., Gurevich, I.B., Martinelli, M., Salvetti, O., Trusova, Y.: Ontology driven
Approach to Cell Image Analysis. In OGRW07, 7th Open German-Russian Workshop, Et-
tlingen, Germany (2007)
7. Colantonio, S., Gurevich, I.B., Martinelli, M., Salvetti, O.,Trusova, Y.: Thesaurus-based Im-
age Analysis Ontology. In SAMT07, 2nd Int. Conf. on Semantic and Digital Media Tech-
nologies - 2007 - 5-7 Dec. Genova, Italy (2007)
8. Dwyer, S.J.: A personalized view of the history of PACS in the USA. In: Proceedings of
the SPIE, Medical Imaging 2000: PACS Design and Evaluation: Engineering and Clinical
Issues”, edited by G. James Blaine and Eliot L. Siegel. Vol. 3980 (2000) 2-9
9. eXist: http://exist.sourceforge.net/ (2007)
10. Gholap, A., Naik, G., Joshi, A., Rao, C.V.K.: Content-Based Tissue Image Mining. In
CSBW’05, IEEE Computational Systems Bioinformatics Conference Workshops (2005)
11. Hunter, J.: Adding Multimedia to the Semantic Web - Building and Applying MPEG-7 On-
tology. Multimedia Content and the Semantic Web: Standards, and Tools, Giorgos Stamou
and Stefanos Kollias (Editors), Wiley (2005)
12. OWL: http://www.w3.org/TR/owl-features/ (2004)
13. Perner, P.: Mining Knowledge in Medical Databases. In Data Mining and Knowledge Dis-
covery: Theory, Tools and Technology. In SPIE Vol. 4057 (2000) 359-369
14. XQuery language: http://www.w3.org/TR/xquery/ (2007)
15. Zhang, J., Hsu, W., Lee M.L.: Image Mining: Issues, Frameworks and Techniques. In
MDM/KDD’2001, the Second International Workshop on Multimedia Data Mining, San
Francisco, CA, USA (2001)
9292
