
A
Practical Partial Parser for Biomedical
Literature Summarization
Yasunori Yamamoto
1
and Toshihisa Takagi
2
1
Department of Computer Science, University of Tokyo, Tokyo, Japan,
2
Department of Computational Biology, University of Tokyo, Kashiwa, Japan
Abstract. We present a partial parser called TeLePaPa (TextLens Par-
tial Parser) to identify subjects and predicate verbs (SPVs) in a sentence
of abstracts of MEDLINE citations. The performance of TeLePaPa is the
precision of 96.7% and 97.1% for the SPV detection, respectively, and
the recall of 91.3% and 94.9%, respectively. We found that there was a
similarity in the distribution of the pairs of SPV over different research
topics in the domain. In addition, we found that the power law holds
for the relationship of the number of citations uncovered by SPV pairs
and its rank. That is, only a half of the pairs covered about 90% of all
the citations. This fact enables us to efficiently scan the huge amount of
biomedical literature.
1 Introduction
Thanks to the rapid development of the information technology, a vast amount
of data can be inexpensively stored and transfered as an electro-magnetic form.
The field of biomedicine is not an exception. The researchers can obtain several
kinds of valuable information on the Internet for free, such as literature (by
PubMed
3
), gene (by LocusLink
4
), and protein (by SwissProt
5
).
In addition, the evolution of the biomedical research technology has enabled
the researchers to get lots of data all at once (i.e., Microarray). As a result, those
cases have happened more than before which not only the genes a researcher
knows well, but also those unfamiliar to him or her relate to the studying phe-
nomenon. In this situation, he or she usually begins to survey those unfamiliar
genes by exploring research papers which discuss them since essential knowledge
of biomedicine is still stored in literature. Major starting point to find those pa-
pers is to conduct a PubMed search, where a user inputs some keywords which
he or she thinks most relevant to the concept in his or her mind. However, since
a PubMed search retrieves related literature from MEDLINE database which
stores more than 12 million citation data, the result is often too many for a
3
h
ttp://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=PubMed
4
http://www.ncbi.nlm.nih.gov/LocusLink/
5
http://www.ebi.ac.uk/swissprot/index.html
Yamamoto Y. and Takagi T. (2004).
A Practical Partial Parser for Biomedical Literature Summarization.
In Proceedings of the 1st International Workshop on Natural Language Understanding and Cognitive Science, pages 75-85
DOI: 10.5220/0002678700750085
Copyright
c
SciTePress

single person to check all of listed papers in a practical period of time. The cost
of learning a new gene is high and all the more if the amount of data to be
surveyed is larger.
One approach to solve this issue is to provide users with a more compre-
hensive literature retrieval system such as ExploreMed [1]. With ExploreMed, a
user can narrow down papers by interacting with the system so as to find only
his or her interesting ones. Another approach, that is our goal, is to retrieve re-
lated literature and summarize it for a user to reduce the learning cost. We are
working on developing such a system called TextLens. This system includes text
retrieval and multi-document summarization. To summarize free text appeared
in biomedical literature, we assume to use several natural language processing
(NLP) approaches. While quite a few efforts have been made for text summa-
rization [2, 3, 4, etc...], few has been done in biomedical domain, especially for
focusing on titles and abstracts (we call the title and the abstract of a paper
“a citation data” hereafter unless otherwise mentioned.). Our short-time goal is
to summarize text appeared in the a citation data obtained from MEDLINE.
This goal is to narrow down our final goal to a single document summarization
system which only focuses on a citation data.
As for the reason why we use only citation data, we assume that they are
compiled by the authors, and that the essence of the paper, that is what we want
to extract, should be included there. Besides, to use full papers for text processing
is difficult in this domain [5]. Obtaining full papers in a form for text processing
is not as easy as getting a citation data from MEDLINE. We need to get a license
from each publisher but some do not permit. Even though we can obtain full
papers, they are usually written in HTML (Hyper Text Mark-up Language) and
have not been standardized yet. For these reasons, we assumed that extracting
relevant sentences from the citation data was enough to get a main idea of a
paper and decided to use MEDLINE for our system. A relevant sentence here
means a sentence which contains information important to a biological researcher
who is seeking the contribution made by the authors of a paper.
In this paper, we introduce a partial parser called TeLePaPa (TextLens Par-
tial Parser) to find a relevant sentence. An input of TeLePaPa is a Part-Of-Speech
(POS) tagged sentence output by the parser FDGLite [6]
6
, and its output is an
annotated sentence indicating the portions of its subjects and predicate verbs
(SPVs)
7
. To cope with long and complex collocations often seen in biomedi-
cal literature, we took an approach of chunking them before passing them to
FDGLite. The chunking method is based on Barrier word method [7]. After get-
ting a FDGLite output, TeLePaPa looks up SPVs. Our approach is a rule based
and deterministic method which iterates a replacement of a POS tag of each
word or chunk (term) with a letter denoting its syntactic characteristics such as
FDGLite is the reduced-functionality version of FDG, a full parser which outputs a
result of its deep analysis of a sentence. We use FDG to compare with TeLePaPa.
Some sentences (e.g., compound sentence) have multiple subjects and/or predicate
verbs.
76

noun phrase or preposition
8
. The iteration ends when no more rules to replace
can be applied. Following the iteration, another set of rules are applied.
We applied TeLePaPa to two corpora of citation data whose research topics
were mainly on MAP-kinase (MAPK, a protein) and Aquaporin (AQP, a human
UV-regulated gene), respectively. The result of TeLePaPa showed its precision
of 96.7% and 97.1% for the SPV detection, respectively, and the recall of 91.3%
and 94.9%, respectively. We compared results of TeLePaPa with those of FDG
and found that TeLePaPa outperformed FDG in the biomedical domain. In
addition, we evaluated the effectiveness of our way of chunking by comparing
the processing time with that of FDG and Charniak parser and confirmed it. An
interesting finding of our experiments is that the power-law (similar to Zipf’s
law [8]) is still effective to the relationship of the numb er of citation data not
covered by SPV pairs and its rank. While a couple of approaches to find a
relevant sentence can be thought such as using a statistical information of terms’
appearances or positions of sentences in an abstract, using the feature is an
efficient way to do it.
In the following sections, we will describe our motivation, the method and
its relating studies, the result, some discussions of our study, and a conclusion.
2 Motivation
Many existing parsers such as FDG, ENGCG [9], and Charniak parser [10] are
focusing on newswire or published books. Text in such media is usually written
by professional writers, and therefore there is rarely a grammatical or syntactic
inappropriateness or any writing off the standard way.
Biomedical papers, on the other hand, are usually written by researchers
who are not professional in writing or have limited English proficiency as me,
and therefore text in those papers is not necessarily well written in terms of
grammatical aspects. Consequently, in order for a computer to appropriately
process biomedical papers, it should be kept in mind that not only the amount
of them is large, but also linguistic “quality” or writing style is different from
paper to paper. This means it is more challenging to process such text.
Furthermore, biomedical literature has some characteristics different from the
other text such as newswire (see Table 1). First, the average number of words in
a sentence is more than that of newswire. Our preliminary experiment showed
that the average number of words in the citation data was 14.4, which was 0.7
more than that of Penn Tree Bank (PTB) corpus [11], 13.7.
Second, more long collocations appear in biomedical literature. This is be-
cause many gene names, protein names, or otherwise any chemical substance
name used in the literature consist of multiple words such as JNK interacting
protein 1 or insulin-like growth factor. Our preliminary experiment showed that
the average number of words of collocations in biomedical literature was 2.9,
which was 0.4 more than that in newswire, 2.5.
Since these syntactically representing letters are really roughly assigned, they are
not necessarily grammatically correct.
77
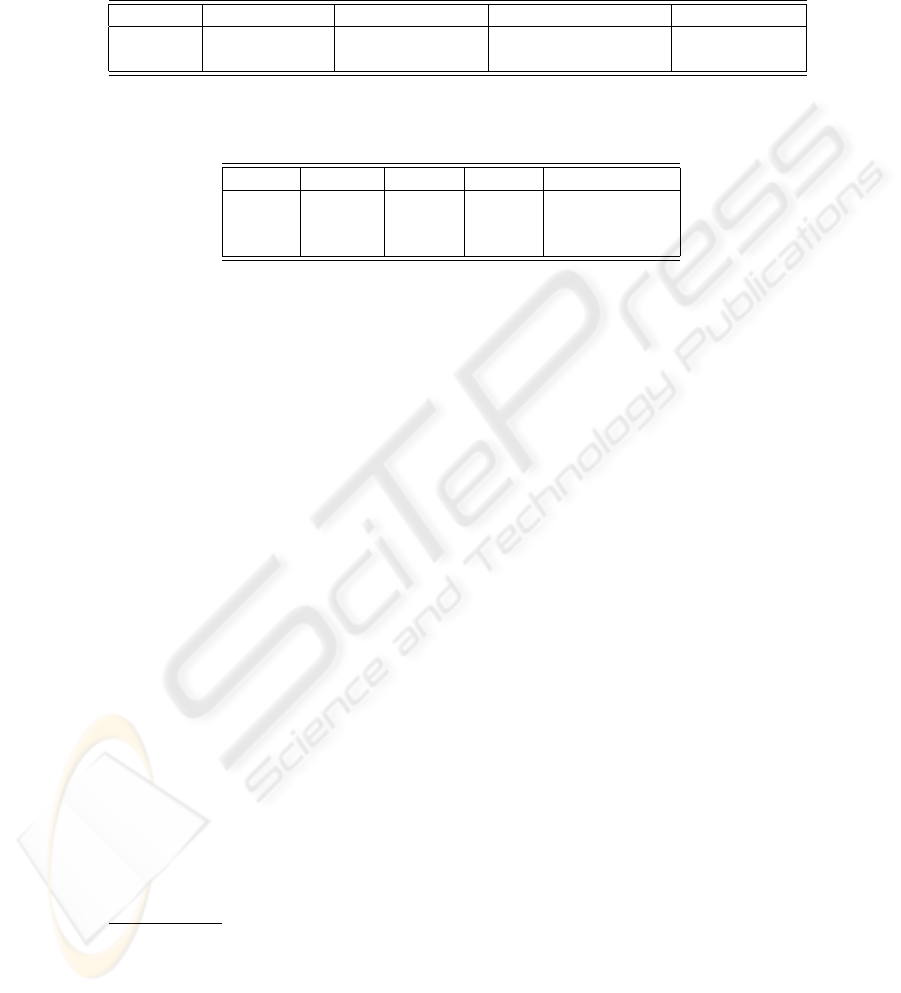
Table 1. Comparison of word distributions (average). We calculated these numbers
on 56,899 sentences of citation data randomly retrieved from MEDLINE, and 52,731
sentences of PTB corpus. Collocations here means those ngrams appear more than
three times in a million ngrams (window size = 5).
# of words/stc.
# of words/colloc.
interval of longer CCs
# of colloc./stc.
MEDLINE
14.4
2.9
11.6
4.6
PTB
13.7
2.5
18.9
1.9
Table 2. Comparison of full parsers. The results of FDG above show the average
numbers of those results evaluated on subjects, objects, and predicate verbs.
FDG
FDG
FDG
Charniak Parser
Precision
93.3
96.3
93.7
91.1 (ave.)
Recall
86.0
92.0
89.0
Target
broadcast
literature
newswire
newswire
Third, more coordinate conjunctions (CCs) which have at least three coor-
dinate terms appear in biomedical literature. This means that expressions such
as “A, B, and C” or “A, B, or C” appear more frequently. Our preliminary
experiment showed that the average interval of the appearance of those CCs in
biomedical literature was 11.6 sentences, which was 7.3 less (i.e., more frequent)
than that in newswire, 18.9.
Another issue is due to existing full parsers. While a parser having an ability
to fully and correctly parse biomedical literature in a practical period of time is
ideal, to develop such a parser is quite difficult. Even though a couple of existing
full parsers perform well for newswire or general books (see Table 2), they per-
form poorly when parsing biomedical literature. Our preliminary experiment on
the full parser FDG showed its accuracy of about slightly more than fifty percent
(54.2%) for 192 randomly retrieved MEDLINE citation data
9
. In addition, full
parsing takes time to take care of the grammatical and syntactical ambiguity.
In this situation, the motivation of our developing a shallow parser is based
on our assumption that a SPV plays a vital role in a sentence for us to catch
an idea of it. Accordingly, identifying those two elements is a first step to get
an idea the authors of a paper want to express. Therefore, while extracting an
entire and precise idea of a sentence from it by deep parsing is ideal if possible,
considering the current research environment, to identify only the two elements
still makes sense in terms of the pragmatic reasons, speed and accuracy. One
way of using TeLePaPa we assume is to search a trigger to extract a relevant
sentence from a vast number of biomedical papers.
Concerning related works, few efforts have been done so far concerning rel-
evant sentence extraction from a citation data of a biomedical paper. As for
9
We evaluated it by checking if it correctly added a dependency function tag (defined
in the user’s manual of FDG) to each word. The result was that 104 sentences were
correctly parsed out of 192.
78
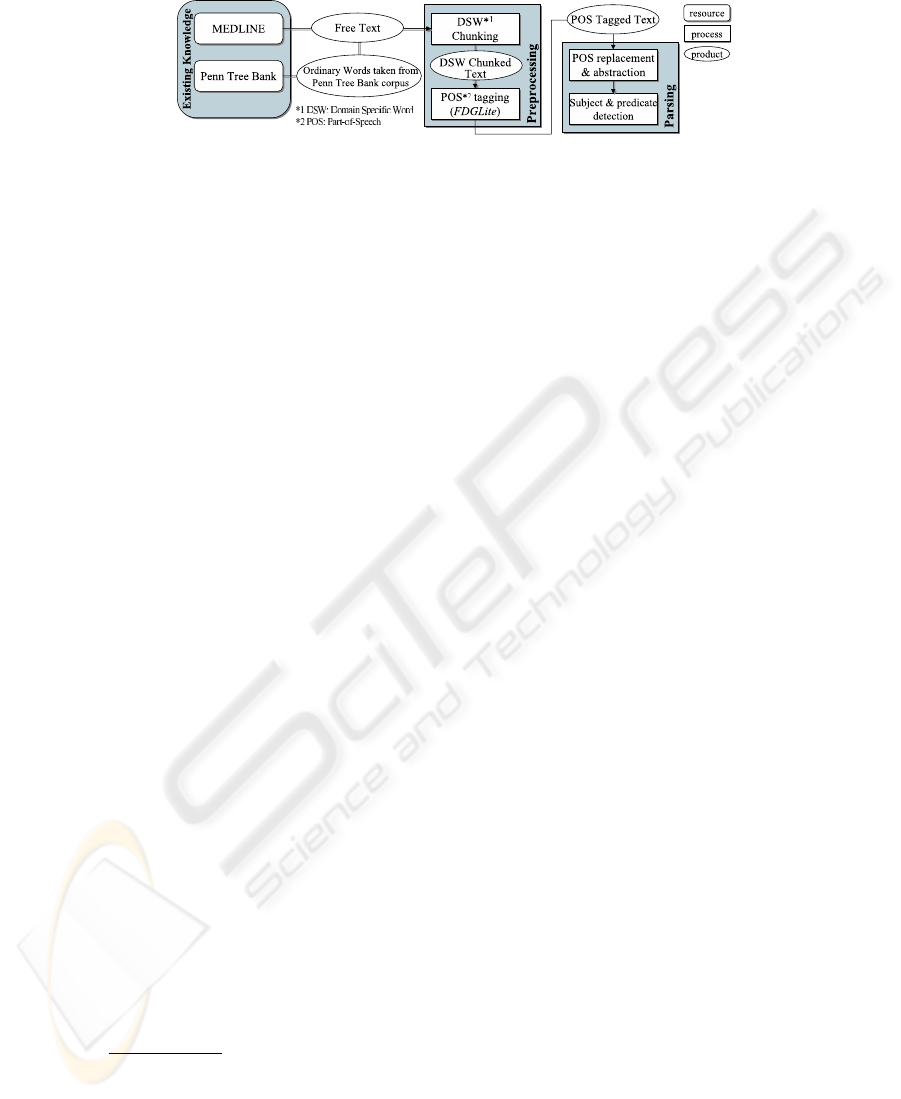
Fig. 1. The system diagram
keyword extraction from a citation data of scientific papers, Hulth made an ef-
fort [12]. It relates to our study in that he claims the citation data is important
to catch a main idea of it.
Another related effort is NLS Java Repository Project
10
, a project of National
Library of Medicine (NLM). They have developed a parser called PhraseX to ex-
tract noun phrases [13]. While correctly extracting noun phrases from biomedical
literature is absolutely challenging and important, TeLePaPa focuses on another
task, that is, to extract SPVs.
Pustejovsky et al. [14] developed a parser to identify and extract biomolecular
relations from biomedical literature. On “inhibition” relations, the parser showed
the precision of 90% and the recall of 57%. While this result demonstrates its p o-
tential for extracting another biomolecular relations, its performance is unknown
when it is applied to general biomedical papers to extract SPVs.
3 Method
Figure 1 shows a brief system diagram of TeLePaPa. The basic way of TeLePaPa’s
processing follows the tradition of partial parsing [15, 16, 17]. It deterministically
analyzes a sentence by applying several rules and replacing some terms with its
abstract expression in a bottom-up manner, but does not recursively apply.
Preprocessing At the beginning of the entire process, text (i.e., citation data)
is taken from MEDLINE database and split into one sentence per one line. This
process is done by a tool developed internally.
After that, domain specific words are to be chunked to minimize any mis-
parsing by FDGLite due to its lack of lexical knowledge of this domain. This is
a solution to the first and second issues discussed in the previous section. We
built a user-made lexicon consisting of those chunks. This process is based on
Barrier word method, whose idea is that frequently appearing words can be used
as delimiters for each domain specific words such as gene name, protein name,
or any technical terms usually not incorporated in a general parser’s lexicon. In
TeLePaPa, the delimiter words are all of those included in PTB corpus.
Following the chunking, POS tagging is done by FDGLite. It adds both
syntactic and morphological tags to each word
11
.
10
http://umlslex.nlm.nih.gov/nlsRepository/doc/userDoc/index.html
11
FDG adds word-form, lemma, syntactic tag, and morphological tags.
79
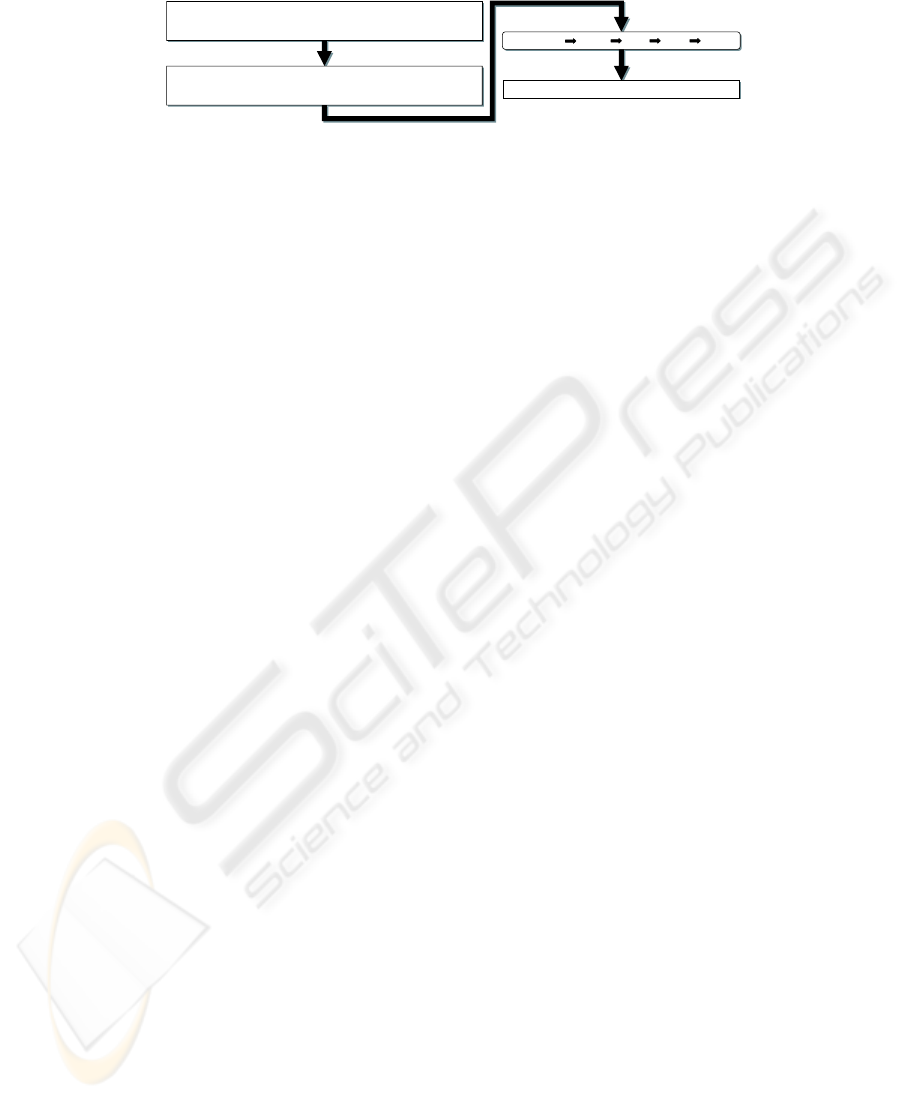
Statistical analyses of 42 cases and 32 controls carrying the codon 31 Arg
allele identified hypertension ( odds ratio , 4.33 ) and family history of cancer
( odds ratio , 2.81 ) as positive risk factors.
Statistical analyses of 42 cases and 32 controls carrying the codon 31 Arg
allele identified hypertension ( odds ratio , 4.33 ) and family history of cancer
( odds ratio , 2.81 ) as positive risk factors.
Statistical analyses / of / 42 cases / and / 32 controls carrying the cod on 31 Arg
allele / identified / hypertension / ( / odds ratio , 4.33 / ) / a
nd / family history /
of / cancer / ( / odds ratio , 2.81 / ) / as / positive risk factors / .
Statistical analyses / of / 42 cases / and / 32 controls carrying the cod
on 31 Arg
allele / identified / hypertension / ( / odds ratio , 4.33 / ) / and / family history /
of / cancer / ( / odds ratio , 2.81 / ) / as / positive risk factors / .
Noun Phrase N, Verb V, ‘of’ 0, ‘as’ 1, ...
N0NCNVNqNcNpCN0NqNcNp1N
N0NCNVNqNcNpCN0NqNcNp1N
Fig. 2. An example of the replacement process.
Parsing TeLePaPa firstly replaces terms with a letter which basically represents
their syntactic function such as noun phrase or verb phrase (see Fig. 2). This pro-
cess goes as follows: 1) make chunks by using the following syntactic constituents
as delimiters: prepositions, conjunctions, punctuations, relatives, infinitives, and
verbs, and 2) replace each chunk with a syntactically representing letter. We call
a product of this process “LS” (Letterized Sentence). Since adverbs, adjectives,
or phrases of these kinds never become a constituent of a subject or predicate
verb, they are needed to be trimmed. In addition, detecting the scope of a CC is
needed to appropriately identify multiple subjects or predicate verbs as seen in
complex sentences. These special cares are a solution to the third issue discussed
in the previous section.
TeLePaPa fulfills these processes by applying a series of rules to a LS. Its
method is deterministic and therefore it can reduce a cost of the process. Each
rule consists of a regular expression and the parser substitutes a portion of a LS
with another at which the expression matches. Followings are some examples of
those rules (expressed in the form of Perl regular expression):
– M[MA]+P -> MP (e.g., will[M] be[M] precisely[A] predicted[P]),
– M+A*V -> V (e.g., could[M] have[M] sharply[A] reduced[V]), or,
– q[^p]+p -> B (e.g., ([q] i.e., five times higher )[p]).
The method to recognize the scop e a CC covers follows DP (Dynamic Program-
ming) algorithm, that is, to scan letters just before and after the conjunction
and to get scores of syntactic similarities of them.
Once getting a LS, then a process of identifying a SPV begins. When con-
sidering the metho d, we assumed that all the sentences in biomedical literature
have a SPV, and that there is no sentence having a form of inversion, that is, an
inversion of the order of a SPV. Of course, this hypothesis is not true realistically,
but such a case rarely happens especially in scientific literature. Furthermore,
since our goal is to extract a key sentence from an abstract by which the authors
want to express their contribution, it is far more rare that such a sentence is
contrary to our assumption.
Consequently, the basic pro cess of this phase is to firstly find an independent
verb (i.e., a predicate verb) and next to find a NP located before the found verb
(a subject). To cope with a compound sentence, the parser previously recognizes
a comma used to connect multiple clauses and iterates the independent verb
search until all the clauses are scanned.
80

4 Result
We developed TeLePaPa on randomly extracted sentences appeared in those
papers which mentioned human UV-regulated genes. Then, we applied it to two
corpora of MEDLINE citation data: a corpus mentioning MAPK (5,312 citations
and 51,696 sentences)and a corpus mainly mentioning AQP (776 citations and
7,916 sentences). Training set and the two corpora used to evaluate TeLePaPa
are mutually excluded. The reason why we took these corpora are their functional
importance in an organism and our internal purposes. We calculated precisions
and recalls for SPV identifications for each corpus. Those values are defined as
follows: 1) Precision is equal to the number of subjects/predicate verbs correctly
identified divided by the number of those which were actually identified, and
2) Recall is equal to the number of subjects/predicate verbs correctly identified
divided by the total number of those which should be identified. Since the number
of sentences were too many for a single person to check all of them, we assessed
the TeLePaPa’s results for the randomly taken 147 sentences of the MAPK and
210 of the AQP to evaluate its performance. In addition, we evaluated the result
of FDG to assess the effectiveness of our method. The FDG parsed the same
sentences of AQP as TeLePaPa did.
Table 3 shows those results. As for the MAPK sentences, both the precision
and the recall for the predicate verb detection (97.1% and 94.9%) are better than
those of the subject detection (87.2% and 89.8%). As for the AQP sentences,
however, whereas the precision of the subject detection was better than that
of the predicate verb detection (96.7% and 94.2%), the recall of the predicate
verb detection was better than that of the subject detection (92.4% and 91.3%).
Compared to the FDG output, the precision of the TeLePaPa’s output was not
as good as it, but the recall was better, especially for the predicate verb detection
(17.5% higher). This result shows that FDG takes a very conservative approach.
To assess the processing time, we implemented TeLePaPa on a Sun Fire 15000
machine and calculated it. To parse the MAPK corpus, the total elapsed time was
15 minutes including the POS tagging. As for FDG, it took 20 minutes. Moreover,
we applied Charniak parser to the same corpus. Contrary to the TeLePaPa’s
process time, it was almost a whole day long (23 hours 57 minutes).
To observe the distributions of SPV pairs, we counted the numbers of the
citation data which a pair of a SPV covers for each corpus. Besides, to see
the pairs’ distribution in general biomedical literature, we made another corpus
consisting of randomly retrieved 52,568 MEDLINE citation data published in
2002. Table 4 shows the ranks of those pairs for each category in the order of
their frequencies. While the two corpora have different research topics from each
other, both have similar distributions. The randomly obtained corpus also shows
the similarity.
We also investigated the relationship between the number of citation data
covered by a pair and its rank. Figure 3 shows those results. The x-axis denotes
the logarithm of the rank. The y-axis denotes the number of citation data not
yet containing any already counted pairs from the top to that rank. From this
analysis, we found that the power-law holds for the relationships. As for the
81
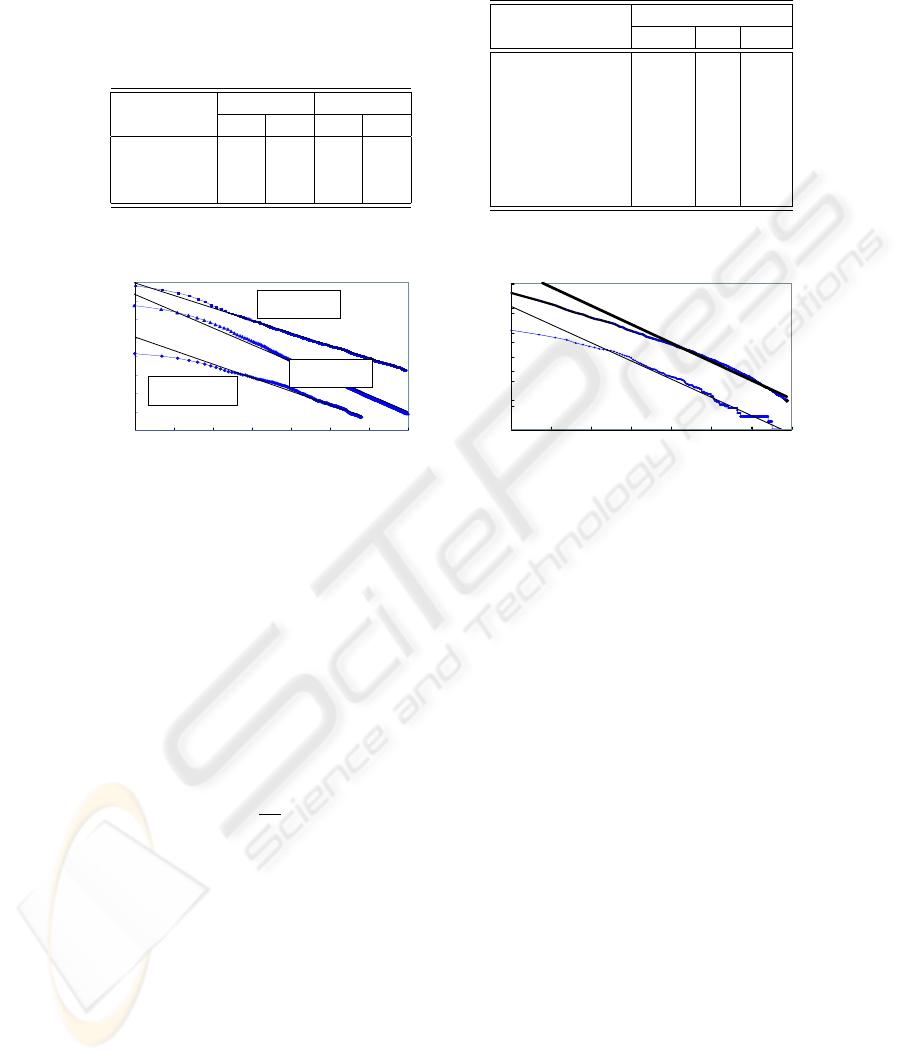
Table 3. Result of TeLePaPa and
FDG. As FDG adds each tag to each
word, we regarded its parsing as good
if a NP containing a subject noun is
appropriately identified.
Precision
Recall
Subj.
Pred.
Subj.
Pred.
MAPK
87.2
97.1
89.8
94.9
AQP
96.7
94.2
91.3
92.4
FDG (AQP)
100
98.8
83.1
74.9
Table 4. The ranks of SPV pairs.
SPV Pair
Rank
MAPK
AQP
RND.
we show
1
1
2
these result suggest
2
4
5
we investigate
3
8
4
we demonstrate
4
-
-
we examine
5
5
6
we report
7
2
1
we found
6
3
3
4.5
5
5.5
6
6.5
7
7.5
8
8.5
0123 456
7
10
10.2
10.4
10.6
10.8
11
y = -0.3669x + 7.0231
R
2
= 0.9828
(left)
Aquaporin
y = -0.3373x + 8.4704
R
2
= 0.9984
(left)
MAP-kinase
y = -0.1142x + 10.919
R
2
= 0.999
(right)
Random
Uncovered Papers (log)
Rank (log)
Fig. 3. The number of citation data
not covered by SPV pairs and the rank
of a pair.
y
=
-
1
.
4947
x
+
10
.
146
R
2
=
0
.
9679
0
2
4
6
8
10
12
0123456
7
U
n
c
o
ve
r
e
d
P
a
p
e
r
s
(
log
)
R
a
n
k
(
log
)
y
=
-
0
.
3133
x
+
8
.
7201
R
2
=
0
.
9771
6
6
.
5
7
7
.
5
8
8
.
5
(
P
r
e
di
ca
t
e
V
e
r
b
, L
e
ft
)
(
S
ubj
ec
t
,
R
igh
t
)
Fig. 4. The number of citation data
not covered by subject or predicate
verb and their rank for “MAPK”.
MAPK pairs, the following relationship holds: log(F ) = 0.3373 log(R)+8.4704,
where F denotes the number of not covered citation data and R denotes a rank
at that number, respectively. Its correlation coefficient is 0.9984, and therefore
those two numbers are highly related with each other. Let N and p
i
be the total
number of the citation data and a set of citation data containing the pair at
its rank of i, respectively, and S means the number of the elements the set
S contains. Since F = N
S
R
i=1
p
i
, the equation above can be expressed as
S
R
i=1
p
i
= N
C
R
α
, where N = 5312, C = 4771, and α = 0.3373, respectively.
As for AQP, N = 776, C = 1122, and α = 0.3669, respectively. In case of
the random corpus, the same power law still holds (the correlation coefficient is
0.999), and N = 52568, C = 55215, and α = 0.1142, resp ectively.
To see whether or not the same characteristic can be found in another func-
tional constituent, we investigated distributions of subjects and predicate verbs,
respectively (Fig. 4). The right and left y-axes denote the number of citation data
not covered by subjects and predicate verbs, respectively. Those correlation co-
efficients are 0.9771 and 0.9679, and therefore both of the two relationships do
not follow the power-law as much as the case of the SPV pairs.
82

5 Discussion
The demand on a precise and practical text processing in biomedical domain is
high. However, due to the nature of the domain, one of the fundamental functions
for it, named entity task has not answered that demand yet [5]. Nomenclature
issue has always been a great worry in this community [18, 19, 20, etc...]. In this
work, we took another approach toward the text processing, that is, rather than
tackling the named entity task at first, grappling the basic sentence structure
detection, which has the same goal.
The drawbacks of TeLePaPa is mainly due to two issues: mal-detection of
complex sentence structures and inappropriate output of FDGLite. The former
issue can be observed when the next sentence as an example is parsed.
Our data shows that in contrast with BC2, DD1 is an inducer of X activation
measured by cell aggregation, chemiluminescence, or release of A4 serotonin.
It is a complex sentence having a that-clause, and that that-clause has several
commas; The first two of them are used to separate the adverbial clause from the
rest, and the others are to connect each constituent of the CC. While appropriate
recognition of the function of a comma at that point is quite difficult, the way of
using the second comma causes more mis-parsing currently. To find the scope of
a that-clause is more difficult if there is a comma after the “that”. Our current
parsing method of using only syntactic information has a limitation, since there
is a case that syntactic expressions or LSes of two different sentences having
a that-clause are exactly the same, but that the scopes of the that-clauses are
different from each other. A solution to cope with this issue without deteriorating
the simplicity should be considered.
As for the other issue, inappropriate FDGLite output, it is because there are
some cases for FDGLite to mis-annotate POS tags in biomedical literature. For
example, the word “assay” is rarely used as a verb in the domain, but FDGLite
tends to regard it as a verb. One way to compensate this problem is to make
another custom lexicon to tell FDGLite the distribution of correct POS tags for
each word frequently appeared in the domain. However, the current version of
FDGLite cannot accept that kind of lexicon.
Despite these issues, TeLePaPa achieved an acceptable performance by em-
ploying a simple approach to tackle rather complex and idiosyncratic sentences in
biomedical literature. The approach includes the in-advance chunking of domain
specific words and the parsing by the regular-expression based deterministic re-
placement. Currently, TeLePaPa only detects SPVs of a sentence, but these two
syntactic elements are fundamental to capturing the idea of the sentence. Even
though there is a case of needing deep analyses, in-advance sentence retrieval by
TeLePaPa makes them more efficient by reducing the total number of sentences
to be deeply analyzed. Our discovery of the power-law in the distribution of SPV
pairs enables it. As for the case of MAPK citation data, the top 500 pairs appear
in about ninety percent (89.1%) of all the 5,312 citation data. Considering the
total number of the pairs appeared in at least three citation data is more than
one thousand (1,024), only a half of them cover almost all the citation data, and
83

those most frequently used pairs seem to be triggers to extract a main idea from
an abstract such as these result suggest or we conclude.
In addition, the distributions of the top pairs have similarities across the two
corpora of citation data which have different research topics from each other.
The corpus of the randomly retrieved citation data also shows the similarity,
suggesting that the characteristic can be seen irrespective of research area in
biomedicine. Furthermore, an analysis of a long, complex sentence can be eased
by splitting it into two parts, before and after the predicate verb since in English
no modification happens between words beyond the predicate verb. For these
reasons, it is expected to encourage better analyses of sentences in the domain
and to lead to the performance improvement of information extraction from the
enormous amount of biomedical literature.
6 Conclusion
By using the series of rules described as regular expressions to deterministically
parse a sentence, TeLePaPa accomplished fast parsing to identify SPVs without
loosing the p erformance. The most significant part of the contribution to the
performance improvement as compared to FDG is to previously chunk domain
specific words particularly used in biomedicine and not covered by general lex-
icons. As a result of taking the approach, TeLePaPa achieved the precision of
96.7% and 97.1% for SPV detection, respectively. It also achieved the recall of
91.3% and 94.9% for the SPV detection, respectively. Subject and predicate verb
detection helps extract a most relevant sentence from an abstract of a citation
data. In addition, we found that the power-law holds for the distribution of the
number of citation data not covered by SPV pairs sorted by the order of the fre-
quency. Our work contributes to more efficient text processing for the biomedical
literature.
Acknowledgments
We thank to Hiroko Ao, Kanebo Ltd. for providing us with an excellent sentence
splitter called JASMINE. We also thank to Asako Koike for reviewing this paper
and giving us invaluable comments. This work is partly supported by the grant
Grant-in aid for scientific research on priority areas (c) Genome Information
Science from the Ministry of Education, Culture, Sports, and Technology, Japan.
References
[1] Perez-Iratxeta, C., Keer, H.S., Bork, P., Andrade, M.A.: computing fuzzy associ-
ations for the analysis of biological literature. Biotechniques 32 (2002) 1380–1385
[2] Barzilay, R., Elhadad, N., McKeown, K.R.: Inferring strategies for sentence or-
dering in multidocument news summarization. Journal of Artificial Intelligence
Research 17 (2002) 35–55
84

[3] Teufel, S., Moens, M.: Summarizing scientific articles: Experiments with relevance
and rhetorical status. Computational Linguistics 28 (2002) 409–446
[4] Teufel, S., Moens, M.: What’s yours and what’s mine: Determining intellectual
attribution in scientific text. In: Proceedings of Joint SIGDAT Conference on
Empirical Methods in NLP and Very Large Corpora. (2000)
[5] Dickman, S.: Tough mining, the challenges of searching the scientific literature.
PLoS Biology 1(2) (2003) 144–147
[6] Tapanainen, P., J¨arvinen, T.: A non-projective dependency parser. In: Proceed-
ings of the 5th Conference on Applied Natural Language Pro cessing. (1997)
[7] Tersmette, K.W.F., Scott, A.F., Moore, G.W., Matheson, N.W., Miller, R.E.:
Barrier word method for detecting molecular biology multiple word terms. In:
Pro ceedings of the 12th Annual Symposium on Computer Applications in Medical
Care. (1988) 207–211
[8] Zipf, G.K.: Human Behavior and The Principle of Least Effort. An Introduction
to Human Ecology. Addison-Wesley Press., Reading, MA (1949)
[9] J¨arvinen, T.: Annotating 200 million words: The bank of english project. In:
Pro ceedings of Coling-94, Vol. I, Kyoto, Japan (1994) 565–568
[10] Charniak, E.: A maximum-entropy-inspired parser. In: Proceedings of NAACL-
2000. (2000)
[11] Marcus, M.P., Santorini, B., Marcinkiewicz, M.A.: Building a large annotated
corpus of english: the penn treebank. Computational Linguistics 19(2) (1994)
313–330
[12] Hulth, A.: Improved automatic keyword extraction given more linguistic knowl-
edge. In: Proceedings of the 2003 Conference on Empirical Methods in Natural
Language Processing. (2003) 216–223
[13] Srinivasan, S., Rindflesch, T.C., Hole, W.T., Aronson, A.R., Mork, J.G.: Finding
umls metathesaurus concepts in medline. In: Proceedings of AMIA Symposium.
(2002) 727–731
[14] Pustejovsky, J., Castafio, J., Zhang, J., Kotecki, M., Cochran, B.: Robust rela-
tional parsing over biomedical literature: Extracting inhibit relations. In: Pro-
ceedings of 2002 the Pacific Symposium on Biocomputing. (2002) 362–373
[15] Hindle, D.: Deterministic parsing of syntactic non-fluencies. In: Proceedings of
the 21st Annual Meeting of the Association for Computational Linguistics. (1983)
123–128
[16] McDonald, D.D.: Robust partial parsing through incremental, multi-algorithm
pro cessing. In Paul, S.J., ed.: Text-Based Intelligent Systems. Lawrence Erlbaum
Asso c (1992) 83–99
[17] Weischedel, R., Meteer, M., Schwartz, R., Ramshaw, L., Palmucci, J.: Coping
with ambiguity and unknown words through probabilistic models. Computational
Linguistics 19(2) (1993) 359–382
[18] Andrade, M.A., Valencia, A.: Automatic annotation for biological sequences by ex-
traction of keywords from medline abstracts. development of a prototype system.
In: Proceedings of International Conference on Intelligent System for Molecular
Biology. (1997) 25–32
[19] Ohta, Y., Yamamoto, Y., Okazaki, T., Uchiyama, I., Takagi, T.: Automatic con-
struction of knowledge base from biological papers. In: Proceedings of Interna-
tional Conference on Intelligent System for Molecular Biology. (1997) 218–225
[20] Fukuda, K., Tsunoda, T., Tamura, A., Takagi, T.: Toward information extraction:
Identifying protein names from biological pap ers. In: Proceedings of 1998 the
Pacific Symposium on Biocomputing. (1998) 707–718
85
