Segmentation of Cell Membrane and Nucleus using Branches
with Different Roles in Deep Neural Network
Tomokazu Murata
1
, Kazuhiro Hotta
1
, Ayako Imanishi
2
, Michiyuki Matsuda
2
and Kenta Terai
2
1
Meijo University, 468-8502, Nagoya, Aichi, Japan
2
Kyoto University, 606-8315, Kyoto, Japan
{ matsuda.michiyuki.2c, terai.kenta.5m} kyoto-u.ac.jp
Keywords: Segmentation, Cell Membrane, Cell Nucleus, Convolutional Neural Network, U-Net and Bioimage.
Abstract: We propose a segmentation method of cell membrane and nucleus by integrating branches with different roles
in a deep neural network. When we use the U-net for segmentation of cell membrane and nucleus, the accuracy
is not sufficient. It may be difficult to classify multi-classes by only one network. Thus, we designed a deep
network with multiple branches that have different roles. We give each branch a role which segments only
cell membrane or nucleus or background, and probability map is generated at each branch. Finally, the
generated probability maps by three branches are fed into the convolution layer to improve the accuracy. The
final convolutional layer calculates the posterior probability by integrating the probability maps of three
branches. Experimental results show that our method improved the segmentation accuracy in comparison with
the U-net.
1 INTRODUCTION
For the development of cell biology, it is important to
understand the state of cells accurately. Currently, the
most accurate way to check the state of cells is human
visual inspection. However, it requires much time and
effort. In addition, the results become subjective.
Therefore, the automation of the process is desired in
the field of cell biology. In this paper, we propose an
automatic segmentation method of cell membrane
and nucleus.
In recent years, deep learning gave high accuracy
in many computer vision tasks (Tang and Wu, 2016,
Tseng, Lin, Hsu, and Huang, 2017, Caesar, Uijlings
and Ferrari, 2016, Ghiasi and Fowlkes, 2016). In
particular, encoder-decorder CNN such as U-net
(Ronneberger, Fischer and Brox, 2015) and Segnet
(Badrinarayanan, Kendall and Cipolla, 2015) are
recent trend of semantic segmentation. The advantage
of U-net is to integrate the features at shallow layers
and at deep layers, and features which are lost by
convolution are used at deeper layers effectively.
When we apply the U-net to segment cell membrane
and nucleus, the accuracy is not sufficient for cell
biologists. Since it is important to know how cell
nucleus is covered by cell membranes, many
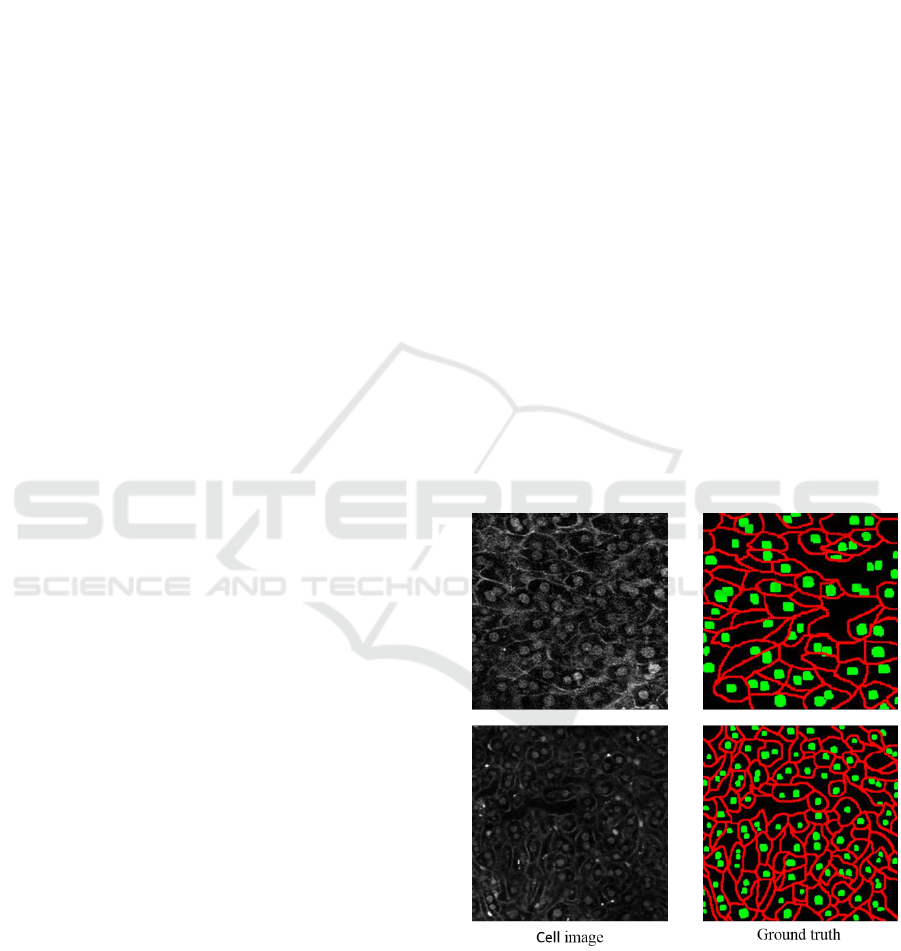
Figure 1: Example of cell images and ground truth labels.
Red shows the cell membrane and green shows cell nucleus.
discontinuities of cell membranes are not good for
biologists. It may be difficult to classify multi-classes
by the standard U-net. In order to address this issue,
we propose to give the U-net multiple branches for
solving different roles.
256
Murata, T., Hotta, K., Imanishi, A., Matsuda, M. and Terai, K.
Segmentation of Cell Membrane and Nucleus using Branches with Different Roles in Deep Neural Network.
DOI: 10.5220/0006717002560261
In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2018) - Volume 4: BIOSIGNALS, pages 256-261
ISBN: 978-989-758-279-0
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
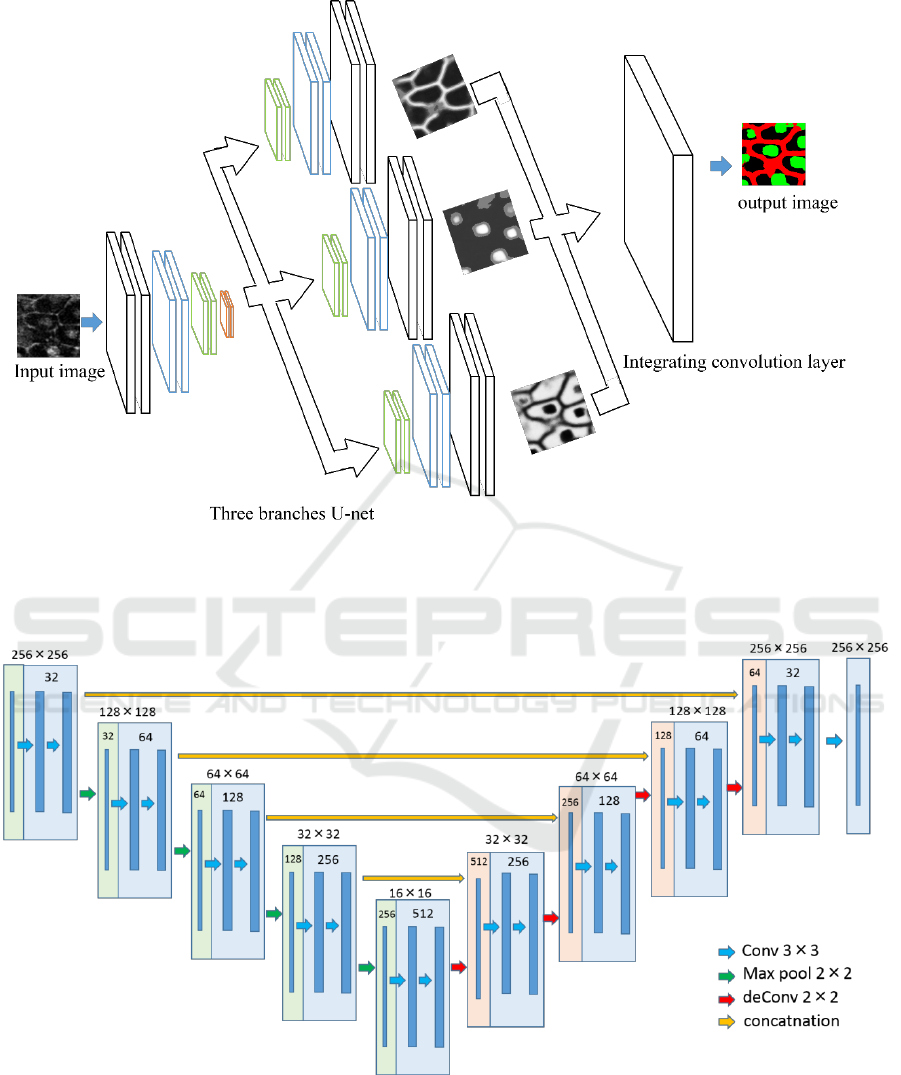
Figure 2: Overview of the proposed network. The decode part is divided into three parts, although all the branches are
connected with the encoder parts and concatenated. This model optimized the output obtained by each branch and the result
of integrating them with soft max cross entropy.
Figure 3: Structure of U-nets used in this paper. The number in the square shows the number of feature map.
In this paper, we use the network structure with
reference to the U-net. The encoded part is the same
structure as the original U-net, and we modified the
decoder part of U-net. The decoder part is divided
into three branches. Each branch has a unique role
that generates a probability map for cell nucleus or
cell membrane or background from the feature maps
obtained by encoder. Finally, single convolution layer
combines the three probability maps, and it gives final
segmentation result.
Segmentation of Cell Membrane and Nucleus using Branches with Different Roles in Deep Neural Network
257
In the experiments, we used 50 fluorescence
images of the liver of transgenic mice that expressed
fluorescent markers on the cell membrane and in the
nucleus. Figure 1 shows the examples of cell image
and ground truth label attached by human experts.
Input for the network is grayscale image and the
output is 3 probability maps that each probability map
is for cell membrane, nucleus and background. Red
shows the cell membrane and green shows cell
nucleus. We evaluated segmentation results by class-
average accuracy, and we confirmed that our
proposed method improved the accuracy of cell
membrane or nucleus in comparison with the U-net.
This paper is organized as follows. In section 2,
we explain the proposed method with different roles.
Dataset and experimental results are shown in section
3. Comparison with conventional U-net is also shown.
Section 4 is for conclusion and future works.
2 PROPOSED METHOD
Figure 2 and 3 show the overview of our proposed
network and the structure of the U-net used in this
paper. Input image is a grayscale image including cell
membrane and nucleus. As described previously, it is
difficult for simple U-net to segment cell membrane
and nucleus simultaneously. Thus, we add branches
to the decoder part of the U-net and assign each
branch to different task. The outputs of three branches
are integrated by a convolution layer to obtain the
final segmentation result. This may be a kind of
curriculum learning (Bengio, 2009). Three decoder
parts try to do only one task, and the final convolution
layer tries to integrate three branches.
At first, an input image is fed into encoder part of
the U-net. Features are extracted by multiple
convolutions and pooling. The encoded feature is fed
into the three decoders with different roles. Each
decoder learns to output the probability map for only
cell membrane or nucleus or background.
Each branching decoder part calculates the
probability of each pixel whether the certain class (e.g.
cell nucleus) in 3 classes or not. Thus, the input of
final convolutional layer is probability maps with 6
channels, and the output of it is the probability map
for 3 classes; cell nucleus, cell membrane and
background. We use softmax cross entropy as the
losses for three branches and final integration layer.
In this paper, we use the weighted sum of losses
of three decoders with different roles and integrated
layer. The whole networks are trained simultaneously.
The weighed loss is defined as
Loss =
+
,
(1)
where c is the class that is one of cell nucleus,
membrane and background.
is the loss generated
by the c-th branch.
is the loss for the
convolutional layer for final segmentation result. In
experiments, we set those parameters as
empirically.
Each loss is defined as
,
(2)
,
(3)
where
,
are the output of three branches and
final convolutional layer respectively. “i” means the
i-th pixel in an input image, and
are the
ground truth.
is class-balancing weight
(Badrinarayanan, Kendall and Cipolla, 2015). Class-
balancing is a method for weighting the loss of each
class according to the number of pixels in each class.
In this paper, background pixels are overwhelmingly
larger than the number of cell nucleus and membranes.
Thus, the network tends to learn to background
dominantly. By applying the weights according to
occurrence of each class, all classes are trained
equally. In this paper, the class weights of cell
membrane, cell nucleus and background are 1, 2.72
and 0.42, respectively.
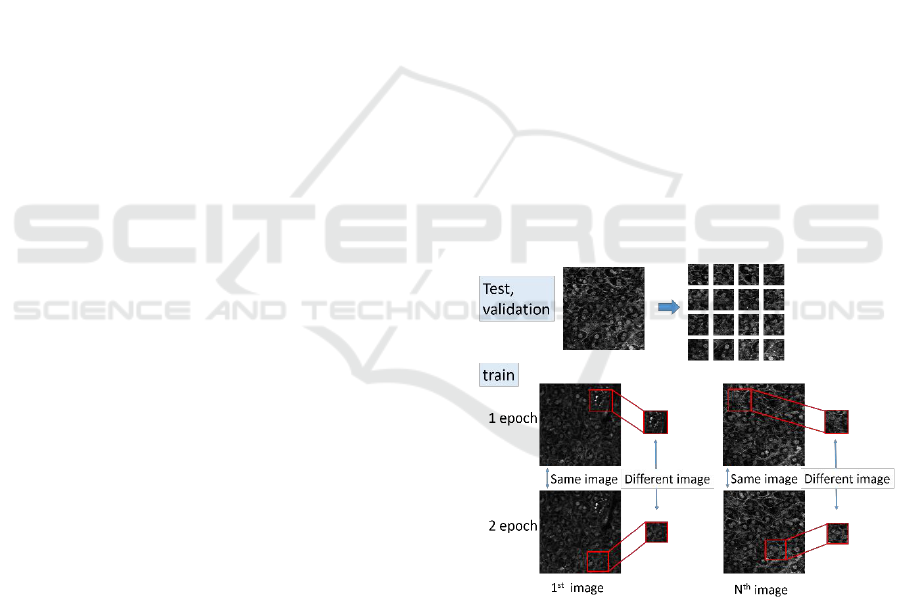
Figure 4: Cropping local region. At every epoch, different
images are cropped from training images. This prevents
overfitting.
3 EXPERIMENTS
This section explains the dataset, evaluation measure
and results. In section 3.1, we describe the dataset
used in this paper. Evaluation measure is also
BIOSIGNALS 2018 - 11th International Conference on Bio-inspired Systems and Signal Processing
258

explained in section 3.1. Experimental results are
shown in section 3.2. Comparison result with the U-
net is also shown.
3.1 Dataset and Evaluation Measure
We use original dataset which includes fluorescence
images of the liver of transgenic mice that expressed
fluorescent markers on the cell membrane and in the
nucleus. To train the segmentation network, we
require fluorescence images with ground truth.
However, creating ground truth labels for cell images
is a labor job for cell biologists. Therefore, the
number of images with ground truth is limited. In this
paper, we have only 50 images. The size of those
images is 256 x 256 pixels. Examples of cell images
and ground truth labels are shown in Figure 1. Red
and green show the cell membrane and nucleus. In the
following experiments, 50 images are divided into
three sets; 35 training images, 5 validation images and
10 test images.
To solve the problem on a small number of images,
data augmentation of training images is used.
Concretely, left-right mirroring and rotations with 90
degrees are combined, and the number of training
images is 8 times larger. In addition, we crop local
regions with 64 x 64 pixels from the augmented
images randomly. Since the size of input images for
the U-net is 256 x 256 pixels, the cropped images are
resized to 256 x 256 pixels and used for training.
To prevent the overfiting, we crop local regions
randomly at each epoch when we train the network.
Figure 4 shows the overview of this process. Since
different local regions with ground truth are cropped
randomly at each epoch from training images, the
network can avoid the overfit.
When we evaluate test images, a test image with
256 x 256 pixels is divided into 4 x 4 without overlap.
The cropped 64 x 64 images are resized to 256 x 256
pixels and fed into the proposed method. By this
processing, the number of images used for the final
test is 160 and the number of validation is 80.
In experiments, we use class average accuracy as
the evaluation measure because the main purpose of
this research is to segment cell membrane and nucleus.
Since the area of background is the largest, pixel-wise
accuracy heavy depends on the accuracy of
background. On the other hand, since class average
accuracy is the average of accuracy of each class, the
accuracy of small area is influenced to the class
average accuracy.
Since the accuracy of deep learning depends on
the random number, we trained the networks three
times and evaluate the average accuracy.
Figure 5: Comparison of output of no weight branches U-
nets. (a) shows a test local region. (b) shows ground truth.
(c) shows the outputs by three branched decoder parts of U-
net in the proposed method. (d) is the output of the network
with the same structure as the proposed method when we
do not give a role branched decoder parts of U-net.
3.2 Evaluation Results
To show the effectiveness of the proposed method
integrating three branches with different roles, we
also evaluate the network with the same structure as
the proposed method as shown in Figure 2. But we
evaluated the proposed method while changing the
value of . One of them is
. Namely,
this optimizes only the cross entropy loss at the final
output. By the comparison with this network, we
understand the effectiveness of training of three
branches with different roles. Of course, the accuracy
of the U-net as shown in Figure 3 is also evaluated.
Table 1 shows the accuracy of each method. As
described previously, we trained each method three
times and average accuracy is evaluated because the
accuracy of networks depends on random number. In
this paper, each pixel in an input image is classified
into three classes; cell membrane, cell nucleus and
background. Table 1 shows that the accuracy changes
slightly depending on the random number.
The mean accuracy of three time evaluation is
shown in Table 2. We see that the accuracy of the
proposed method outperformed with the U-net.
We also evaluate the network with the same
structure as the proposed method and without giving
a role to the branches. We see that the accuracy is
worse than our proposed method.
Figure 5 shows the outputs of three branches in
the proposed method and those in the network
without specific roles. (a) and (b) show a test local
region and its ground truth label. (c) and (d) show the
outputs of the branched decoder part in both methods.
Segmentation of Cell Membrane and Nucleus using Branches with Different Roles in Deep Neural Network
259
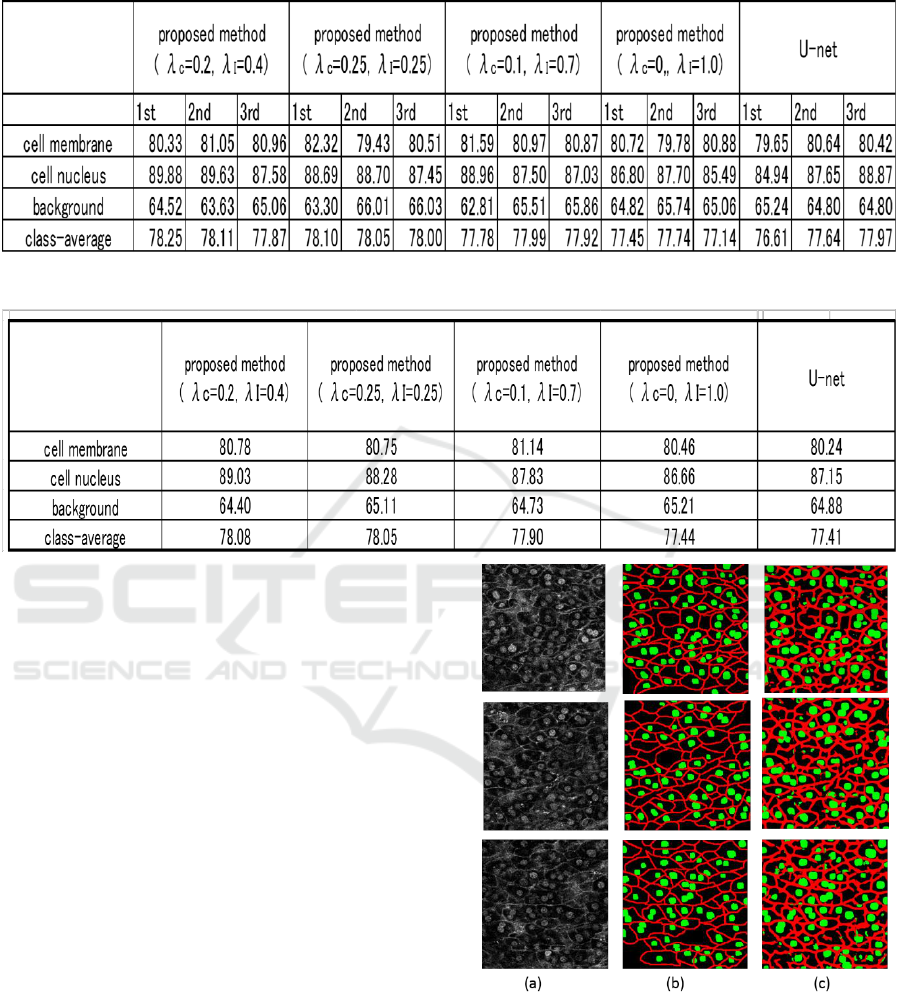
Table 1: Accuracy at three times evaluation.
Table 2: Average accuracy of three times evaluation.
The proposed method gave obviously better maps
than the network without a specific role. This result
demonstrated that the integration of networks with
specific role is effective to improve the accuracy.
(d) is the result that we trained the network without
calculating the losses at the three branches. We
conducted experiments three times under the same
conditions, but similar results are obtained that one of
the three branches had the function of focusing on the
segment of the cell nucleus. In this paper, we give
each clear role to three branches, but we found that
this network has a little ability to share the roles
automatically.
Finally, we show the segmentation results by the
proposed method in Figure 6. Figure 6 (a) and (b)
show the test images and their ground truth labels. (c)
shows the results by the proposed method. We see
that overall segmentation is good though the cell
membrane is conspicuous. Figure 7 shows the
segmentation results of local regions by the proposed
method (
= 0.2,
= 0.4) and the U-net.
Figure (a) and (b) are the test local regions and
their ground truth labels. (c) is the results by the
proposed method. (d) is the results by the U-net. We
see that the segmentation accuracy of cell membrane
and nucleus is improved in comparison with the U-
net.
Figure 6: (a) shows test images. (b) shows ground truth. (c)
shows the results by the proposed method.
In the results shown in the first and second row,
there is a case that cell membranes disconnected by
U-net are connected by the proposed method. The
effectiveness of branches with different roles is
demonstrated by experiments.
BIOSIGNALS 2018 - 11th International Conference on Bio-inspired Systems and Signal Processing
260
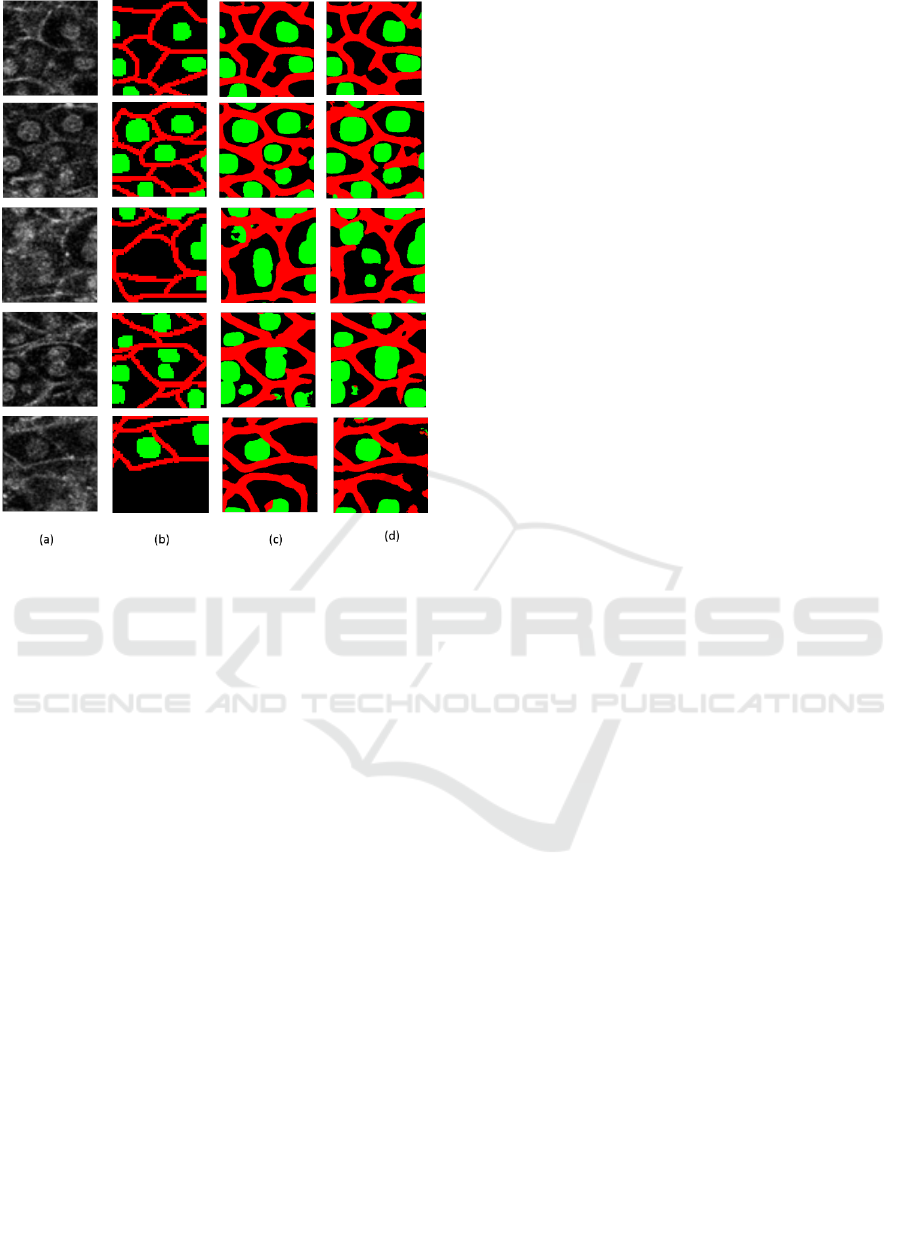
Figure 7: comparison results of local regions. (a) shows test
regions. (b) shows ground truth. (c) shows the results by the
proposed method using three branches. (d) shows the
results by the U-net.
4 CONCLUSIONS
We improved the segmentation accuracy by using
branches with different roles and final convolution
layer. Three branches segment only cell membrane or
nucleus or background, and the final convolution
layer for integrating the outputs of three branches
estimate the posterior probability of each pixel. By
assigning each branched decoder to a different role,
the accuracy was improved.
We crop a local region with 64 x 64 pixels from a
test image without overlap, and the output of the local
region is put to final segmentation result. If we apply
the proposed method to local regions with
overlapping manner, some segmentation results are
obtained at the same pixel. The integration of those
results will improve the accuracy further. It is a
subject for future works.
ACKNOWLEDGEMENTS
This work is partially supported by MEXT/JSPS
KAKENHI Grant Number 16H01435”Resonace Bio”.
REFERENCES
Ronneberger, O., Fischer, P., and Brox, T., 2015. U-net:
Convolutional networks for biomedical image
segmentation. In International Conference on Medical
Image Computing and Computer-Assisted
Intervention (pp. 234-241). Springer, Cham.
Tang, Y., and Wu, X., 2016. Saliency detection via
combining region-level and pixel-level predictions with
cnns. In European Conference on Computer Vision (pp.
809-825). Springer International Publishing.
Tseng, K. L., Lin, Y. L., Hsu, W., and Huang, C. Y., 2017.
Joint Sequence Learning and Cross-Modality
Convolution for 3D Biomedical Segmentation. arXiv
preprint arXiv:1704.07754.
Caesar, H., Uijlings, J., and Ferrari, V., 2016. Region-based
semantic segmentation with end-to-end training.
In European Conference on Computer Vision (pp. 381-
397). Springer International Publishing.
Ghiasi, G., and Fowlkes, C. C., 2016. Laplacian pyramid
reconstruction and refinement for semantic
segmentation. In European Conference on Computer
Vision (pp. 519-534). Springer International
Publishing.
Badrinarayanan, V., Kendall, A., and Cipolla, R., 2015.
Segnet: A deep convolutional encoder-decoder
architecture for image segmentation. arXiv preprint
arXiv:1511.00561.
Bengio, Y., 2009. Learning deep architectures for
AI. Foundations and trends in Machine Learning, 2(1),
1-127.
Segmentation of Cell Membrane and Nucleus using Branches with Different Roles in Deep Neural Network
261
