
Symmetric Generative Methods and tSNE: A Short Survey
Rodolphe Priam
University of Southampton, University Road, Southampton SO17 1BJ, U.K.
Keywords:
Data Visualization, Generative Model, Latent Variables, tSNE, Survey.
Abstract:
In data visualization, a family of methods is dedicated to the symmetric numerical matrices which contain
the distances or similarities between high-dimensional data vectors. The method t-Distributed Stochastic
Neighbor Embedding and its variants lead to competitive nonlinear embeddings which are able to reveal the
natural classes. For comparisons, it is surveyed the recent probabilistic and model-based alternative methods
from the literature (LargeVis, Glove, Latent Space Position Model, probabilistic Correspondence Analysis,
Stochastic Block Model) for nonlinear embedding via low dimensional positions.
1 INTRODUCTION
In visualization of high-dimensional data, the obser-
vations in the available sample are vectorial: the rows
or the columns of a numerical data matrix or table.
An extensive literature exists and diverse approaches
have been developped until today in this domain of
research. Among the existing methods, t-Distributed
Stochastic Neighbor Embedding (t-SNE or tSNE)
(van der Maaten and Hinton, 2008) is a recent method
which improves the previously proposed one called
Stochastic Neighbor Embedding (SNE) (Hinton and
Roweis, 2003). The idea of the SNE is to introduce
soft-max probabilities which transform the matrix of
distances defined in multidimensional scaling (MDS)
(Sammon, 1969; Chen and Buja, 2009) into vectors
of probabilities in order to deal with a Kullback-
Liebler divergence instead of an Euclidean distance.
As a dramatic improvement in comparison to former
researches on distance matrices for visualization,
tSNE is widely used today in various domains in
order to proceed to data analysis. For instance, this
method helps the researchers to improve any data
processing which asks for an effective reduction or
to proceed to the data analysis itself by looking at
a synthetic view (Mahfouz et al., 2015; Shen et al.,
2015; Delauney et al., 2016; Chen et al., 2017).
Nextafter, this introduction presents further the
notations, the purpose and the way it is achieved for
a survey on related probabilistic methods.
Data: Let’s have the available high-dimensional
data as a set of data vectors in a space with M dimen-
sions as follows:
X = {x
i
∈ R
M
;1 ≤ i ≤ N}.
Let define W = (w
i j
) from the distance between x
i
and x
j
, for instance d
i j
=k x
i
− x
j
k, with w
i j
= d
i j
,
or more generally a function of d
i j
. Typically, a weig-
hted nearest neighbors graph such as a heatmap with
for instance w
i j
= e
−d
i j
/τ
for τ > 0 when d
i j
is enough
small, and w
i j
= 0 otherwise. A weighted graph
G = (V,E, W ) is defined from V , E, and W which
stands respectively for the set of vertices (i or v
i
), ed-
ges (e
i j
or (i, j)), and weights w
i j
. An edge e
i j
comes
from a pair of vertices (v
i
, v
j
) in the graph of nearest
neighbors. It is also denoted
¯
E for the set of pairs of
vertices that are not neighbors.
Reduced Vectors: The purpose is to summarize
X by finding relevant lower dimensional representati-
ons:
Y = {y
i
∈ R
S
;1 ≤ i ≤ N}.
Generally S = 2 as the visualization appears in the two
dimensional plane, even if three dimensions or even
more remain possible. The paper is interested on the
modeling of low dimensional positions via their pai-
rwise distances/similarities plus bias/intercept terms.
The parameterization involved is as follows,
y
T
i
˜
y
j
+ b
i
+
˜
b
j
or k y
i
−
˜
y
j
k
2
or δ
i j
= δ(y
i
,
˜
y
j
). (1)
where δ(.,.) is a function of a distance or similarity.
Note that
˜
y
i
and y
i
are not always chosen equal. In the
Euclidean case, it can be rewritten the latent terms as
follows,
b
i
+
˜
b
j
+ y
T
i
˜
y
j
=
˜
b
i
+
˜
˜
b
j
−
1
2
ky
i
−
˜
y
j
k
2
. (2)
This parameterization can be interpreted as descri-
bing latent variables in a low-dimensional Euclidean
space, for the two forms just above. Some other
356
Priam, R.
Symmetric Generative Methods and tSNE: A Short Survey.
DOI: 10.5220/0006684303560363
In Proceedings of the 13th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2018) - Volume 3: IVAPP, pages
356-363
ISBN: 978-989-758-289-9
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
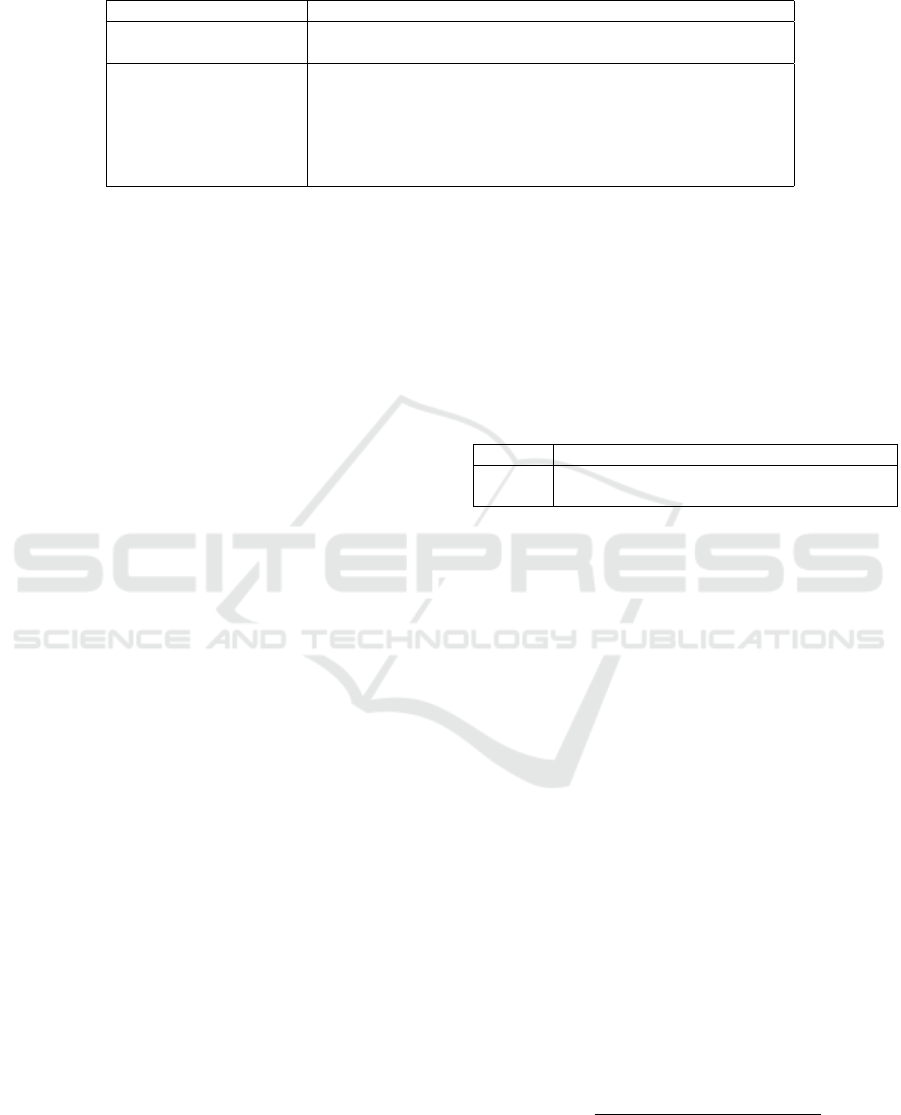
Table 1: Methods presented (third row) in the survey with the modeling in stake for δ
i j
. The column named Probabilistic
means that the method has or has not a generative/model-based foundation with a probabilistic density/mass function.
Method name Probabilistic Modeling Penalization Input
MDS no Euclidean no X , W
Laplacian Eigenmap no Inner product no X , W
SNE, tSNE, ... no Divergence yes (implicit) X , W
LargeVis yes Bernoulli, weights yes (explicit) X , W
Glove yes Log-linear no X (or W )
Probabilistic CA yes Poisson no X (or W )
LSPM/LCPM yes GLM no W
SBM (re-parameterized) yes GLM no W
methods for visualization such as LLE, KPCA,
GPLVM are already compared in (Lee and Verleysen,
2007) for instance and are not presented in this
survey paper as they seem to not follow this para-
meterization. The methods such as linear principal
component analysis (PCA) or factor analysis (FA)
with the embedding (Cunningham and Ghahramani,
2015) of the type k x
i
− Ωy
i
k
2
with Ω a loading
matrix, are not detailed neither. Methods with an
embedding of the type k y
i
− y
(k)
k
2
where y
(k)
is the
position of a cluster as in Parametric Embedding (PE)
(Iwata et al., 2007), or Probabilistic Latent Semantic
Visualization (PLSV) (Iwata et al., 2008) are not
detailed but briefly discussed at subsection 3.5. In the
following sections it is considered only methods with
the parameterization given in (1) or (2).
Objective Function: For finding suitable values
of Y one can look for having d
i j
and δ
i j
enough simi-
lar (large or small) according to a relevant criterion.
The general form of the modeling of the whole set of
methods presented in this survey extends the former
MDS having for objective function,
∑
i j
(d
i j
− δ
i j
)
2
.
It can be added the criterion
∑
i j
w
i j
δ
i j
from Lapla-
cian Eigenmap (Belkin and Niyogi, 2003) (see also in
clustering, Spectral Clustering (von Luxburg, 2007))
as listed in Table 1. The general form is either a me-
asure of distance or gap between functions of d
i j
and
δ
i j
for non probabilistic approaches, or either a like-
lihood from a probabilistic model with an indepen-
dence hypothesis and the parameters depending on
δ
i j
for modeling w, a random variable from W . In
the following, most of the methods include the con-
struction of a vicinity graph with edges w
i j
before mo-
deling the mapping otherwise such a graph is assumed
to be already available.
Illustration: As an example, the dataset in (Gi-
rolami, 2001) is visualized with the data X and W
where the later is for ten nearest neighbors and the
weights are equal to 1 for any observed edge and
to 0 otherwise. The visualization from a) CA+X
(Benzecri, 1980), b) CA+W , c) tSNE+X (van der
Maaten and Hinton, 2008), (d) LargeVis+W (Tang
et al., 2016) and (e) Kruskal’s non-metric MDS+X
are compared in Table 2. The considered indicators
are the average of the Silhouettes (Rousseeuw, 1987)
denoted S-Index and the Davies-Bouldin index (Da-
vies and Bouldin, 1979) denoted DB-Index.
Table 2: Indicators for comparing the quality of projection
from the five methods with the dataset of 1000 binarized
images of handwritten digits with 10 classes.
(a) (b) (c) (d) (e)
S-Index 0.01 0.43 0.50 0.51 0.03
DB-Index 5.82 1.69 0.97 1.34 2.29
If the visual map from CA+W is clearly better
than from CA+X , MDS performs between both. The
two other methods lead to better separated frontiers
for visualizing the natural classes but a graph with
only binary weights is used for CA and LargeVis in
this example.
The following sections present tSNE with its ap-
proximations and its variants, the recent generative
methods for visualization and the perspectives.
2 tSNE, APPROXIMATIONS AND
VARIANTS
In this section, tSNE is briefly presented and its vari-
ants for faster training or enhanced modeling.
2.1 t-Distributed Stochastic Neighbor
Embedding
The method tSNE (van der Maaten and Hinton, 2008)
minimizes the divergence between two discrete distri-
butions. It is first defined:
p
j|i
=
exp(−d(x
i
,x
j
)
2
/2σ
2
i
)
∑
j
0
6= j
exp(−d(x
i
,x
j
0
)
2
/2σ
2
i
)
.
Each parameter σ
i
is set such as the perplexity
2
−
∑
j
p
j|i
log p
j|i
of the conditional distribution P
i
=
Symmetric Generative Methods and tSNE: A Short Survey
357

(p
j|i
) is equal to a positive value smaller than 50 given
by the user. These values are critical for accentuating
the frontiers between the natural classes, as observed
in Spectral Clustering where alternative computations
are available. In former approaches SNE (Hinton and
Roweis, 2003) and NCA (Goldberger et al., 2005)
the probabilities P
i
are directly used in the objective
function. In tSNE it is considered a symmetric distri-
bution P as follows (with also p
i|i
= 0):
p
i j
=
p
j|i
+ p
i| j
2N
.
A distribution Q is defined with t-Student distributi-
ons (instead of the Gaussian ones in SNE/NCA), as
follows (with also q
ii
= 0):
q
i j
=
(1 + ky
i
− y
j
k
2
)
−1
∑
k6=l
(1 + ky
k
− y
l
)k
2
)
−1
.
This solves for the ”crowding effect” where the pro-
jections of the classes are not well separated. The low
dimensional positions y
i
are finally found by mini-
mizing the Kullback-Leibler divergence between the
two distributions P and Q, with the nonlinear and non-
convex objective function,
C(Y ) = D
KL
(P||Q) =
∑
i6= j
p
i j
log
p
i j
q
i j
.
2.2 Approximations
Several approximations have been introduced in the
literature in order to accelerate the computation of the
solution of tSNE -which has by default a quadratic
complexity for the size N- while keeping nearly opti-
mal values for Y . In (van der Maaten, 2013; van der
Maaten, 2014) a sparse approximation computes the
quantities p
j|i
only for N
i
, the nearest neighbors of
x
i
, and keeps them null otherwise. A N-body simu-
lation and thus the Barnes-Hut (BH) algorithm (Ap-
pel, 1985) associated to fast nearest neighbors sear-
ches is proposed in (van der Maaten, 2014) in order
to lower the complexity to only O(N log N) for the
method BH-tSNE via tree-based procedures. See also
(Kim et al., 2016) for a brief overview of the techni-
cal details. Interesting findings have been proposed
recently in the literature in order to accelerate the trai-
ning (Pezzotti et al., 2016; Parviainen, 2016; Kim
et al., 2016) even further. A-tSNE (Pezzotti et al.,
2017) improves the BH-tSNE algorithm by genera-
ting a relevant visual map before a full learning via the
progressive visual analytics paradigm and approxima-
ted nearest neighbors training. The user can choose
to improve some areas of the projection for steerabi-
lity. Improvement or explaination of the training pro-
cedure by alternatives to the sequential approach in-
troduced for tSNE can be found in (Nam et al., 2004;
Yang et al., 2015). Next subsection presents the ex-
tensions of tSNE (and SNE) for improving the mo-
deling foundations.
2.3 Variants, Extensions
Several methods propose to manage the case where
several maps or several datasets are modeled. This
is mainly treated in the literature via weighted sums
for the probabilities q
i j
. More precisely, they are cal-
led multiple maps (van der Maaten and Hinton, 2012;
van der Maaten et al., 2012; Zhang et al., 2013; Xu
et al., 2014), multiple view maps (Xie et al., 2011),
hierarchical maps (Lee et al., 2015; Pezzotti et al.,
2016). Variants of tSNE deal with time series and
temporal data (Rauber et al., 2016) or graph layout
(Kruiger et al., 2017) with a repulsive term as an ad-
ditional penalization.
Other methods aim at improving tSNE (or SNE)
by changing the divergence or the function in the soft-
max of Q or eventually P. The Heavy-tailed Symme-
tric Stochastic Neighbor Embedding (HSSNE) (Yang
et al., 2009) is a generalization of tSNE. Instead of
the t-Student distribution it considers any other heavy-
tailed distribution or any monotonically decreasing
function. In (van der Maaten, 2009) it is proposed
the t-Student distribution with ν degrees of freedom
to optimize jointly with Y . For a spherical embed-
ding, the Euclidean distance is replaced by an inner
product, hence a von Mises-Fisher (vMF) distribution
is considered in vMF-SNE (Wang and Wang, 2016).
Spherical embeddings are also met in two interesting
alternative variants (Lunga and Ersoy, 2013; Lu et al.,
2016).
tSNE and SNE minimize Kullback-Leibler diver-
gences w.r.t. Y , hence alternative divergences (Bas-
seville, 2013) are possible for better robustness. A
weighted mixture of two KL divergences is preferred
in the method Neighborhood retrieval and visualiza-
tion (NeRV) (Venna et al., 2010). In (Lee et al., 2013),
it is proposed a different mixture of KL divergences,
a scaled version of the generalized Jensen-Shannon
divergence. In (Lee et al., 2015) it is proposed an ad-
ditional improvement via multi-scale similarities. In
(Yang et al., 2014) it is proposed the weighted sym-
metric stochastic neighbor embedding (ws-SNE) with
a connection between several divergences (β−, γ− ,
α− and R
´
enyi-divergences). In (Narayan et al., 2015)
it is presented a variant named AB-SNE with a α-β
divergence. In (Bunte et al., 2012) it is developped
a systematic comparison of many divergence measu-
res and shown that no divergence is really better for
simulated noises, the best one needs to be chosen ac-
cording to each dataset. Considering models of diver-
IVAPP 2018 - International Conference on Information Visualization Theory and Applications
358

gence with a generative setting (Dikmen et al., 2015)
for the selection of the optimal extra parameters of
these divergences has also been proposed in (Amid
et al., 2015).
Nextafter, the presented models have an embed-
ding of the positions y
i
in their parameters, with dif-
ferent forms of objective functions.
3 PROBABILISTIC MODELS
In this section, the methods are defined via a proba-
bilistic and model-based foundation. They are ge-
nerally dedicated to the visualization/reduction of a
graph which is constructed from vectorial data.
3.1 Probabilistic CA
Correspondence Analysis (CA) (Benzecri, 1980; Le-
bart et al., 1998) is a matricial method which is ex-
tensively studied for visualizing the rows and the
columns of contingency tables. To perform CA
on a two-way table, the correspondence matrix is
F = X/x.. while x.. stands for its grand total, r =
(r
1
,...,r
N
) for the row margins, and c = (c
1
,...,c
M
)
for the column margins. A low-rank approximation
leads to
b
F =
b
X/x
..
where
b
X is the approximation of X
which can be rewritten according to a reconstruction
formula from the eigen vectors/values of a particular
matrix. By rewritting elementwise (Beh, 2004) this
matricial approximation, this leads to a Poisson ap-
proximation, a probabilistic version of CA:
x
i j
∼ P
x
..
r
i
c
j
1 + y
T
i
˜
y
j
,
or from the approximation
1
at the first order of the
exponential function,
x
i j
∼ P
x
..
e
y
T
i
˜
y
j
+logr
i
+logc
j
.
This also explains why CA can be seen as not fully
linear but this suggests also that from a graph ma-
trix with cells proportional to w
i j
(or p
i j
from tSNE)
instead of x
i j
, a better visualization is expected.
3.2 Glove
Glove (Pennington et al., 2014) is based on a log-
bilinear regression model in order to learn a vector
space representation of words. The weighted regres-
sion for this model can be written as follows:
C(Y ) =
∑
i, j
h(x
i j
)(y
T
i
˜
y
j
+ b
i
+
˜
b
j
− logx
i j
)
2
=
∑
i, j
h(x
i j
)(
˘
y
T
i
˘
˜
y
j
− logx
i j
)
2
.
1
It has also been proposed in the literature to alter the
soft-max with alternative polynomial expressions but not
for visualization.
The function h(.) removes noisy cells in the criterion,
it is equal to (x
i j
/100)
3/4
for x
i j
< 100 and equal to
1 otherwise. For the second line above, it is also de-
noted
˘
y
i
= (b
i
,1,y
T
i
)
T
and
˘
˜
y
i
= (1,
˜
b
i
,
˜
y
T
i
)
T
such as
it is recognized a weighted factorization of the matrix
with cell values logx
i j
, except that a component of the
reductions is constrained to the value 1. This leads to
the approximation,
x
i j
≈ e
y
T
i
˜
y
j
+b
i
+
˜
b
j
,
such as Glove can be seen as a weighted version of
CA, solved via a constrained factorization: a variant
of MDS or Isomap (Tenenbaum et al., 2000) with par-
ticular weights. The original paper (Pennington et al.,
2014) explains how to construct the matrix (x
i j
) from
raw textual data but any symmetric matrix (w
i j
) may
be used for visualization. In (Hashimoto et al., 2016)
it is introduced a fully generative model, based on a
negative binomial distribution NegBin, such that:
x
i j
∼ NegBin
θ,
θ
θ + e
−||y
i
−
˜
y
j
||
2
+b
i
+
˜
b
j
,
where θ controls the contribution of large x
i j
as an
alternative to the function h(.).
3.3 Latent Space Position Models
In data analysis, the models named latent space posi-
tion models (LSPM) are based on a parameterization
of the generalized linear models as seen in (Hoff et al.,
2002) in the binary case. In contrast to the other met-
hods presented herein, the determistic parameters y
i
,
b
i
and
˜
b
j
are replaced by random variables with same
notation in the current subsection. In the latent cluster
position models (LCPM) the positions y
i
are modeled
with a Gaussian mixture as a prior for their clustering
(Handcock et al., 2007).
For LCPM, let denote v a p × N × N array of co-
variates with v
i j
a p-dimensional vector of covaria-
tes for (i, j), β the p-dimensional vector of covari-
ate coefficients, y
i
the S-dimensional position vec-
tor of i, b = (b
1
,··· ,b
N
) the vector of sender ef-
fects,
˜
b = (
˜
b
1
,··· ,
˜
b
N
) the vector of receiver effects,
and h(.) a link function. This leads to the likelihood
function of the observed network w = (w
i j
) as fol-
lows:
f (w|θ,v) =
∏
i, j
f (w
i j
|h
−1
(η
i j
)).
For a complete bayesian model, b
i
and
˜
b
j
are Gaus-
sian with means 0 and respective variances σ
2
b
and σ
2
˜
b
.
It is written:
η
i j
= v
>
i j
β + δ(y
i
,y
j
) + b
i
+
˜
b
j
.
Symmetric Generative Methods and tSNE: A Short Survey
359

The clustering is modeled through a mixture mo-
del with G components where each one is a S-
dimensional Gaussian with mean µ
k
, spherical vari-
ance with parameter σ
2
k
and mixing probability λ
k
,
y
i
∼
∑
G
g=1
λ
k
G
S
(µ
k
,σ
2
k
I
d
).
Alternative distributions such as a t-Student one seem
not have been tested yet for this model. For binary
graphes, a Bernoulli distribution is usually conside-
red for the modeling with h(.) a logit function. The
estimation of Y appears difficult because of the prior
and posterior distributions which are highly nonlinear.
Latent space models are able to find the natural clas-
ses and to select their number according to the best
fit. They can facilitate principled visualization in a
probabilistic setting. Theory on these models can be
found in (Rastelli et al., 2016) and faster inference in
(Raftery et al., 2012) even if the application of these
models may be confined to moderated sizes of graphs
for the current available implementations. The next
method can be seen as a regularized LSPM with an
efficient implementation and no bayesian priors.
3.4 LargeVis
LargeVis (Tang et al., 2016) introduces a probabilis-
tic parametric model dedicated to the visualization
of a nearest neighbors graph. The model is defined
for not only the set of the nearest neighbors but also
all the furthest ones which are forced into a nega-
tive interaction. It is closely related to the previous
approach LINE (Tang et al., 2015b) (see also (Cao
et al., 2015) for an alternative matrix-based learning
and (Tang et al., 2015a) for a semi-supervised variant)
which is non generative. A likelihood for the graph G
can then be written as follows,
L(Y ) =
∏
(i, j)∈E
p(e
i j
= 1)
w
i j
∏
(i, j)∈
¯
E
(1 − p(e
i j
= 1))
γ
.
Where,
P(e
i j
= 1) = g(||y
i
− y
j
||
2
),
stands for the probability that a edge e
i j
exists bet-
ween v
i
and v
j
. For the function g(.), it can be chosen,
g(τ) = 1/(1 + aτ) or g(τ) = 1/(1 + exp(τ)).
while γ is a common weight for the non neighbor
vectices. For a spherical version with g(y
T
i
y
j
), a vMF
distribution function induces an embedding over a sp-
here as in subsection 2.3. LargeVis recalls a Bernoulli
distribution which has been altered for improving its
properties of separability of the clusters by adding the
weigths and the penalization. Concerning this mo-
deling, two remarks are proposed :
- Let’s have ¯w =
∑
(a,b)∈E
w
ab
, ˜p
i j
= w
i j
/ ¯w,
˜
γ = γ/ ¯w,
˜q
i j
= p(e
i j
= 1). By rewriting L(.), it is obtained
a new function to minimize w.r.t. Y ,
`
γ
(Y )
= −
logL(Y )
¯w
+
∑
(i, j)∈E
˜p
i j
log ˜p
i j
=
∑
(i, j)∈E
˜p
i j
log
˜p
i j
˜q
i j
−
˜
γ
∑
(i, j)∈
¯
E
log(1 − ˜q
i j
).
Hence, it is recognized a criterion in two parts.
The term on the left side is roughly similar to the
criterion of tSNE as a divergence but without ( ˜q
i j
)
normalized to sum to 1 and without t-Student dis-
tributions. The other term on the right side is for
a penalization. They insure the local and global
projections respectively.
- It seems appealing to try to improve other distri-
butions such as a Poisson one which may replace
the first term,
P(e
i j
= w
i j
) = p(e
i j
= 1)
w
i j
,
to get eventually an expressive quantity for the
non observed edges with P(e
i j
= 0). A probabilis-
tic interpretation -when w
i j
is an integer- remains
the product between the likelihood of the obser-
ved edges with the likelihood of the non observed
edges with a weighting for regulating the impor-
tance of each one:
L
P
(Y ) =
∏
(i, j)∈E
(
˜
δ
i j
)
w
i j
e
−
˜
δ
i j
w
i j
!
∏
(i, j)∈
¯
E
e
−γ
˜
δ
i j
.
Here
˜
δ is the result from a function of the quantity
δ
i j
such as the exponential one. In this alterna-
tive likelihood, the weighting is modeled explici-
tely and the model is fully generative. The term
to the left with a Poisson mass distribution for E
recalls LSPM without bayesian priors. When in
˜
δ
i j
an exponential function is chosen, the term to
the right for
¯
E recalls the penalization in Elastic
Embedding (Carreira-Perpi
˜
nan, 2010), except the
weighting. Additional weighting is via the para-
meters with respectively,
˜
δ
i j
= e
δ
i j
+logα
for E and
˜
δ
i j
= e
δ
i j
+logw
i j
for
¯
E. With α > 0, this paramete-
rization may lead to a weighting similar to Elastic
Embedding for the non observed edges.
3.5 Stochastic Block Model
As presented in (Matias and Robin, 2014; Daudin
et al., 2008) the stochastic block model (SBM) is de-
fined for a random graph on a set V = {1,...,N} of
N nodes (i or v
i
). Let’s have z = {z
1
,...,z
N
} stands
for N independent and identically distributed (i.i.d.)
IVAPP 2018 - International Conference on Information Visualization Theory and Applications
360

discrete hidden random variables with possible va-
lues in {1,...,K}. Let’s have f
γ
(·;γ
z
i
z
j
) a conditio-
nal density function or mass distribution with para-
meter γ = (γ
k`
)
1≤k,`≤K
. The variables w
i j
are random,
i.i.d. conditionally to (z
i
,z
j
) and aggregated into the
random variable w = (w
i j
)
(i, j)∈E
with a given distri-
bution. SBM has for conditional likelihood the follo-
wing expression, where z
i
and z
j
needs to be integra-
ted out by adding their distribution,
f (w|z) =
∏
(i, j)∈E
f (w
i j
|z
i
,z
j
)
=
∏
(i, j)∈E
f
γ
(w
i j
|γ
z
i
z
j
).
The links between the edges and the structure of the
network are sometimes explained by covariates: v
i
at
the node level as in (Tallberg, 2004) via a multino-
mial probit model for the membership of the verti-
ces or v
i j
at the edge level as in (Mariadassou et al.,
2010) via a regression term within the expectations.
This modeling concerns mainly the clustering part
of the stochastic block model which needs to be re-
parameterized for inducing a nonlinear visualization
(if a posteriori methods such as Parametric Embed-
ding (Iwata et al., 2007) are not used). For visualiza-
tion purposes with this model, further parameters can
be embedded. This results into adding the N latent
variables y
i
via δ(y
i
,y
j
) or its corresponding cluster
version δ(y
(k)
,y
(`)
) with eventual bias terms (b
i
and
˜
b
j
or the cluster versions b
(k)
and
˜
b
(`)
). In the bi-
nary case when g(.) is the sigmoidal function, f
γ
can
be written with a Bernoulli mass distribution function
with parameters,
γ
k`
= g(k y
(k)
− y
(`)
k
2
).
This re-parameterization of SBM recall LSPM but
with a different clustering framework as the mixture
model is not a prior but directly introduced in the data
modeling.
A limitation of the approaches above may be seen
in diagonal co-clustering (Tjhi and Chen, 2006): this
suggests that the quantities γ
z
i
z
j
could not be fully free
parameters. The extra parameters for the visualiza-
tion needs to be added in the posterior probabilities
for instance. This leads in the variational EM for the
inference of SBM (with common parameters or not)
to consider the probability that a datum belongs to a
cluster as one of the following expression:
Q
y
i
(z
i
= k;γ) ∝ e
−ky
i
−y
(k)
k
2
as in PLSV,
Q
y
i
(z
i
= k;γ) =
∑
k
0
h
kk
0
τ
ik
0
as in SOM.
For the later case at the bottom, such algorithm in-
troduces the quantities τ
ik
0
as the free parameters and
h
kk
0
as a smoothing matrix from the neighboor no-
des in the Kohonen’s network or self-organizing maps
(SOM) (Kohonen, 1997).
4 CONCLUSION AND
PERSPECTIVES
In this survey, it is proposed an unified overview of
the literature on data visualization with tSNE and with
the recent alternative symmetric generative methods
2
depending on bivariate latent positions. Several links
between these methods are explained for helping the
comparisons of their objective functions. These com-
parisons suggest eventual variants of several existing
methods such as: CA estimated approximatively via
Glove for large matrices, LSPM or SBM regularized
via a probabilistic penalization for the non observed
edges, or the visualization with the mixture models
extended to symmetric matrices via SBM for a sym-
metric self-organizing map for instance, as future ap-
pealing perspectives.
ACKNOWLEDGEMENTS
The author would like to thanks the reviewers for the
valuable comments.
REFERENCES
Amid, E., Dikmen, O., , and Oja, E. (2015). Optimizing
the information retrieval trade-off in data visualization
using α-divergence. ArXiv e-prints.
Appel, A. W. (1985). An efficient program for many-body
simulation. SIAM Journal on Scientific and Statistical
Computing, 6(1):85–103.
Basseville, M. (2013). Review: Divergence measures for
statistical data processing-An annotated bibliography.
Signal Process., 93(4):621–633.
Beh, E. J. (2004). Simple correspondence analysis: A bi-
bliographic review. International Statistical Review,
72(2):257–284.
Belkin, M. and Niyogi, P. (2003). Laplacian eigenmaps
for dimensionality reduction and data representation.
Neural Compututation, 15(6):1373–1396.
Benzecri, J. P. (1980). L’analyse des donn
´
ees tome 1 et 2 :
l’analyse des correspondances. Paris:Dunod.
Bunte, K., Haase, S., Biehl, M., and Villmann, T. (2012).
Stochastic neighbor embedding (SNE) for dimension
reduction and visualization using arbitrary divergen-
ces. Neurocomputing, 90:23–45.
2
The presented methods have very different training al-
gorithms according to their targeted domain (datamining,
data analysis, data visualization or machine learning) and
the size of the datasets their are experimented with. Most
of these methods can ask for diverse estimation algorithms
such as bayesian inference or online gradient optimization
with efficient data structure, efficient memory management,
in order to infer in the faster way Y .
Symmetric Generative Methods and tSNE: A Short Survey
361

Cao, S., Lu, W., and Xu, Q. (2015). GraRep: Learning
graph representations with global structural informa-
tion. In Proceedings of the 24th ACM International
on Conference on Information and Knowledge Mana-
gement, CIKM’15, pages 891–900.
Carreira-Perpi
˜
nan, M. A. (2010). The elastic embedding al-
gorithm for dimensionality reduction. In Proceedings
of the 27th International Conference on Machine Le-
arning, ICML’10, pages 167–174.
Chen, L. and Buja, A. (2009). Local multidimensional
scaling for nonlinear dimension reduction, graph dra-
wing, and proximity analysis. Journal of the American
Statistical Association, 104(485):209–219.
Chen, V., Paisley, J., and Lu, X. (2017). Revealing com-
mon disease mechanisms shared by tumors of diffe-
rent tissues of origin through semantic representation
of genomic alterations and topic modeling. BMC Ge-
nomics, 18(Suppl 2):105.
Cunningham, J. P. and Ghahramani, Z. (2015). Linear di-
mensionality reduction: Survey, insights, and gene-
ralizations. Journal of Machine Learning Research,
16:2859–2900.
Daudin, J. J., Picard, F., and Robin, S. (2008). A mixture
model for random graphs. Statistics and Computing,
18(2):173–183.
Davies, D. L. and Bouldin, D. W. (1979). A cluster separa-
tion measure. IEEE Transactions on Pattern Analysis
and Machine Intelligence, 1(2):224–227.
Delauney, C., Baskiotis, N., and Guigue, V. (2016). Tra-
jectory bayesian indexing: The airport ground traffic
case. In IEEE 19th International Conference on In-
telligent Transportation Systems (ITSC), pages 1047–
1052.
Dikmen, O., Yang, Z., and Oja, E. (2015). Learning the in-
formation divergence. IEEE Transactions on Pattern
Analysis and Machine Intelligence, 37(7):1442–1454.
Girolami, M. (2001). The topographic organization and vi-
sualization of binary data using multivariate-bernoulli
latent variable models. IEEE Transactions on Neural
Networks, 12(6):1367–1374.
Goldberger, J., Hinton, G. E., Roweis, S. T., and Salakhut-
dinov, R. R. (2005). Neighbourhood components ana-
lysis. In Advances in Neural Information Processing
Systems 17, pages 513–520. MIT Press.
Handcock, M. S., Raftery, A. E. and Tantrum, J. M. (2007).
Model-based clustering for social networks. JRSS-A,
170(2), 301–354.
Hashimoto, T. B., Alvarez-Melis, D., and Jaakkola, T. S.
(2016). Word embeddings as metric recovery in se-
mantic spaces. TACL, 4:273–286.
Hinton, G. E. and Roweis, S. T. (2003). Stochastic neig-
hbor embedding. In Advances in Neural Information
Processing Systems 15, pages 857–864. MIT Press.
Hoff, P. D., Raftery, A. E., and Handcock, M. S. (2002).
Latent space approaches to social network analysis.
Journal of the American Statistical Association, The-
ory and Methods, 97(460).
Iwata, T., Saito, K., Ueda, N., Stromsten, S., Griffiths,
T. L., and Tenenbaum, J. B. (2007). Parametric em-
bedding for class visualization. Neural Computation,
19(9):2536–2556.
Iwata, T., Yamada, T., and Ueda, N. (2008). Probabilis-
tic latent semantic visualization: topic model for vi-
sualizing documents. In Proceeding of the 14th ACM
SIGKDD international conference on Knowledge dis-
covery and data mining, KDD’08, pages 363–371.
Kim, M., Choi, M., Lee, S., Tang, J., Park, H., and Choo, J.
(2016). PixelSNE: Visualizing Fast with Just Enough
Precision via Pixel-Aligned Stochastic Neighbor Em-
bedding. ArXiv e-prints.
Kohonen, T. (1997). Self-organizing maps. Springer.
Kruiger, J. F., Rauber, P. E., Martins, R. M., Kerren, A.,
Kobourov, S., and Telea, A. C. (2017). Graph Layouts
by t-SNE. Comput. Graph. Forum (Proc. of EuroVis),
36(3):283–294.
Lebart, L., Salem, A., and Berry, L. (1998). Exploring Tex-
tual Data. Springer.
Lee, J. A., Peluffo-Ordonez, D. H., and Verleysen, M.
(2015). Multi-scale similarities in stochastic neig-
hbour embedding: Reducing dimensionality while
preserving both local and global structure. Neurocom-
puting, 169:246–261.
Lee, J. A., Renard, E., Bernard, G., Dupont, P., and Verley-
sen, M. (2013). Type 1 and 2 mixtures of Kullback-
Leibler divergences as cost functions in dimensiona-
lity reduction based on similarity preservation. Neu-
rocomputing, 112:92–108.
Lee, J. A. and Verleysen, M. (2007). Nonlinear Dimensio-
nality Reduction. Springer, 1st edition.
Lu, Y., Yang, Z., and Corander, J. (2016). Doubly Stochas-
tic Neighbor Embedding on Spheres. ArXiv e-prints.
Lunga, D. and Ersoy, O. (2013). Spherical stochastic
neighbor embedding of hyperspectral data. IEEE
Transactions on Geoscience and Remote Sensing,
51(2):857–871.
Mahfouz, A., van de Giessen, M., van der Maaten, L., Huis-
man, S., Reinders, M., Hawrylycz, M. J., and Lelie-
veldt, B. P. (2015). Visualizing the spatial gene ex-
pression organization in the brain through non-linear
similarity embeddings. Methods, 73:79–89.
Mariadassou, M., Robin, S., and Vacher, C. (2010). Unco-
vering latent structure in valued graphs: A variational
approach. Ann. Appl. Stat., 4(2):715–742.
Matias, C. and Robin, S. (2014). Modeling heterogeneity
in random graphs through latent space models: a se-
lective review. ESAIM: Proceedings, 47:55–74.
Nam, K., Je, H., and Choi, S. (2004). Fast stochastic neig-
hbor embedding: a trust-region algorithm. In IEEE In-
ternational Joint Conference on Neural Networks (IJ-
CNN), volume 1, page 123–128.
Narayan, K., Punjani, A., and Abbeel, P. (2015). Alpha-
beta divergences discover micro and macro structures
in data. In Proceedings of the 32nd International Con-
ference on Machine Learning, ICML’15, pages 796–
804.
Parviainen, E. (2016). A graph-based N-body approxima-
tion with application to stochastic neighbor embed-
ding. Neural Networks, 75:1–11.
IVAPP 2018 - International Conference on Information Visualization Theory and Applications
362

Pennington, J., Socher, R., and Manning, C. D. (2014).
Glove: Global vectors for word representation.
In Proceedings of the 2014 Conference on Em-
pirical Methods in Natural Language Processing,
EMNLP’14, pages 1532–1543.
Pezzotti, N., H
¨
ollt, T., Lelieveldt, B. P. F., Eisemann, E., and
Vilanova, A. (2016). Hierarchical stochastic neighbor
embedding. Comput. Graph. Forum (Proc. of Euro-
Vis), 35(3):21–30.
Pezzotti, N., Lelieveldt, B. P. F., van der Maaten, L, H
¨
ollt,
T.,Eisemann, E., and Vilanova, A. (2017). Approx-
imated and User Steerable tSNE for Progressive Vi-
sual Analytics. IEEE Transactions on Visualization
and Computer Graphics, 23(7):1739–1752.
Raftery, A. E., Niu, X., Hoff, P. D., and Yeung, K. Y.
(2012). Fast inference for the latent space network
model using a case-control approximate likelihood.
Journal of Computational and Graphical Statistics,
21(4):901–919.
Rastelli, R., Friel, N., and Raftery, A. E. (2016). Properties
of latent variable network models. Network Science,
4(4):407–432.
Rauber, P. E., Falc
˜
ao, A. X., and Telea, A. C. (2016). Visu-
alizing time-dependent data using dynamic t-SNE. In
Proceedings of the Eurographics / IEEE VGTC Con-
ference on Visualization: Short Papers, EuroVis’16,
pages 73–77.
Rousseeuw, P. (1987). Silhouettes: a graphical aid to the in-
terpretation and validation of cluster analysis. J. Com-
put. Appl. Math., 20:53–65.
Sammon, J. (1969). A nonlinear mapping for data struc-
ture analysis. IEEE Transactions on Computers, C-
18(5):401–409.
Shen, F., Shen, C., Shi, Q., van den Hengel, A., Tang,
Z., and Shen, H. T. (2015). Hashing on nonlinear
manifolds. IEEE Transactions on Image Processing,
24(6):1839–1851.
Tallberg, C. (2004). A bayesian approach to modeling sto-
chastic blockstructures with covariates. The Journal
of Mathematical Sociology, 29(1):1–23.
Tang, J., Liu, J., Zhang, M., and Mei, Q. (2016). Visualizing
large-scale and high-dimensional data. In WWW’16.
Tang, J., Qu, M., and Mei, Q. (2015a). PTE: Pre-
dictive text embedding through large-scale heteroge-
neous text networks. In Proceedings of the 21th
ACM SIGKDD International Conference on Know-
ledge Discovery and Data Mining, KDD’15, pages
1165–1174.
Tang, J., Qu, M., Wang, M., Zhang, M., Yan, J., and Mei,
Q. (2015b). LINE: Large-scale information network
embedding. In Proceedings of the 24th Internatio-
nal Conference on World Wide Web, WWW’15, pages
1067–1077.
Tenenbaum, J. B., de Silva, V., and Langford, J. C. (2000).
A Global Geometric Framework for Nonlinear Di-
mensionality Reduction. Science, 290(5500):2319–
2323.
Tjhi, W.-C. and Chen, L. (2006). A partitioning based algo-
rithm to fuzzy co-cluster documents and words. Pat-
tern Recogn. Lett., 27(3):151–159.
van der Maaten, L. (2009). Learning a parametric embed-
ding by preserving local structure. In Proceedings of
the Twelfth International Conference on Artificial In-
telligence and Statistics (AISTATS’09), volume 5, pa-
ges 384–391.
van der Maaten, L. (2013). Barnes-Hut-SNE. In Procee-
dings of International Conference on Learning Repre-
sentations.
van der Maaten, L. (2014). Accelerating t-SNE using tree-
based algorithms. Journal of Machine Learning Rese-
arch, 15:3221–3245.
van der Maaten, L. and Hinton, G. (2008). Visualizing Data
using t-SNE. Journal of Machine Learning Research,
9:2579–2605.
van der Maaten, L. and Hinton, G. (2012). Visualizing non-
metric similarities in multiple maps. Machine Lear-
ning, 87(1):33–55.
van der Maaten, L., Schmidtlein, S., and Mahecha, M. D.
(2012). Analyzing floristic inventories with multiple
maps. Ecological Informatics, 9:1–10.
Venna, J., Peltonen, J., Nybo, K., Aidos, H., and Kaski, S.
(2010). Information retrieval perspective to nonlinear
dimensionality reduction for data visualization. Jour-
nal of Machine Learning Research, 11:451–490.
von Luxburg, U. (2007). A tutorial on spectral clustering.
Statistics and Computing, 17(4):395–416.
Wang, M. and Wang, D. (2016). vMF-SNE: Embedding for
spherical data. In ICASSP’16, Shanghai, China.
Xie, B., Mu, Y., Tao, D., and Huang, K. (2011). m-SNE:
Multiview stochastic neighbor embedding. IEEE
Transactions on Systems, Man, and Cybernetics, Part
B (Cybernetics), 41(4):1088–1096.
Xu, W., Jiang, X., Hu, X., and Li, G. (2014). Visualization
of genetic disease-phenotype similarities by multiple
maps t-SNE with Laplacian regularization. BMC Med.
Genomics, 7:1–9.
Yang, Z., King, I., Xu, Z., and Oja, E. (2009). Heavy-tailed
symmetric stochastic neighbor embedding. In Advan-
ces in Neural Information Processing Systems 22, pa-
ges 2169–2177. Curran Associates.
Yang, Z., Peltonen, J., and Kaski, S. (2014). Optimization
equivalence of divergences improves neighbor embed-
ding. In Proceedings of the 31st International Confe-
rence on on Machine Learning, ICML’14, pages 460–
468.
Yang, Z., Peltonen, J., and Kaski, S. (2015). Majorization-
minimization for manifold embedding. In Procee-
dings of the Eighteenth International Conference on
Artificial Intelligence and Statistics (AISTATS’15), vo-
lume 38, pages 1088-1097.
Zhang, L., Zhang, L., Tao, D., and Huang, X. (2013). A
modified stochastic neighbor embedding for multi-
feature dimension reduction of remote sensing ima-
ges. ISPRS Journal of Photogrammetry and Remote
Sensing, 83:30–39.
Symmetric Generative Methods and tSNE: A Short Survey
363
