
Microfluidic Platform for Aptamer based Fluorimetric Analysis of
Analytes
Tanu Bhardwaj and Sandeep Kumar Jha*
Centre for Biomedical Engineering, Indian Institute of Technology Delhi, New Delhi-110016, India
Keywords: PMMA, Microfluidics, Aptamer, Biosensing, FAM, Fluorescence.
Abstract: In this work, we are reporting fabrication of a simple and low cost setup for fluorescence detection based on
aptamer probes. For this reason, we fabricated a PMMA-PMMA microfluidic chip using easily available
laboratory techniques and combined the chip with a simple fluorescence detection setup using optical fiber,
filter, detector and a commercial spectroscopy software. In this new approach, we used two different
strategies to use aptamers as probe. In first strategy, detection of any nucleic acid could be targeted using
simple DNA hybridization with aptamer probe. Such strategy can be used in analysis of samples with
specific nucleic acid sequence, such as pathogen. We proved this using known sequence of ssDNA aptamer
probe immobilized on detection zone on microchip and its FAM labeled complementary strand was passed
over it using microfluidic condition. In other strategy, we attempted detection of any protein or biomarker
using sandwich fluorimetric technique with primary and labeled secondary aptamer immobilized on sensing
region. For this, we used thrombin as model target to validate our setup. Both the strategies proved
satisfactory on our setup. Even more, LOD was also impressive. In future, this setup could further be
miniaturized by using a small on-chip CCD array detector, microcontroller based electronics and LabVIEW
software based control.
1 INTRODUCTION
Aptamers are single stranded nucleic acids, DNA or
RNA, which selectively and specifically bind to a
target molecule such as nucleic acids, proteins, cells,
microorganisms, etc. interacting via weak molecular
forces. They often complement antibodies in terms
of binding efficiency and specificity, yet are more
stable and cheaper to produce. Aptamers are picked
for a particular target from a process called SELEX
(Systematic evolution of ligands by exponential
enrichment) which is comparatively simple process
compared to typical antibody production using
hybridoma technique. In addition, host animal is also
not required for the production of aptamers. Extra
resistance against denaturation and ease of chemical
modifications make aptamers friendlier to use in
biosensing (Tennico, 2010; Song, 2012).
Meanwhile, microfluidics has replaced many
analytical and biomedical techniques due to its
reduced size, cost and reagents utilization. Aptamers
and their application in microfluidics are inspiring
researchers to create a bridge between two
drastically different fields of biological and
analytical techniques. Aptamers with microfluidics
have already been used for analysis of biological
targets/analytes like thrombin, VEG-165, peptides,
cancer cells, C-reactive protein, viruses, microbes
and various other proteins or biomarkers (Xu, 2010).
Out of which, viruses, pathogens or microbes are
identified by these aptamers due to their specific
microbial proteins, lipopolysaccharides or nucleic
acids. On the other hand, biomarkers (proteins)
originating in the case of cancer allow their detection
using aptamers (Viscidi, 1987; Su, 2015).
Detection of various diseases associated with
microbes was made possible by simple hybridization
of aptamer probe labelled with dye to the
complementary nucleic acid strand from the microbe
(Tennico, 2010). Besides, aptamers and
hybridization principle has been used for DNA
microarrays, single nucleotide polymorphism
detection, gene expression studies and nucleic acid
diagnostic applications (Wang, 2011; Abu-Salah,
2015). Furthermore, various proteins and biomarkers
have been identified using primary and dye labeled
secondary aptamers sandwich assay like ELISA
technique. The use of fluorescent dyes in such
218
Bhardwaj, T. and Jha, S.
Microfluidic Platform for Aptamer based Fluorimetric Analysis of Analytes.
DOI: 10.5220/0006645002180223
In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2018) - Volume 1: BIODEVICES, pages 218-223
ISBN: 978-989-758-277-6
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
sandwich protocol required sophisticated
instrumentation including a microscope (Tennico,
2010; Fenzyl, 2016). Hence, in our work, we
developed a simple setup for fluorescence detection
using PMMA-PMMA microfluidic chip and
aptamers as probe.
The choice of polymethyl methacrylate (PMMA)
slides as a substrate was due to its sturdiness and
possibility for immobilization of aptamer as a probe.
PMMA is also a choice in fabrication of
microfluidics chips because of its low cost and
optical clarity (Tennico, 2010). Various techniques
are available for fabrication of microfluidic chips
like lithography, micromachining, laser ablation,
thermal embossing etc. While, in this work, we used
simple techniques like laser ablation for cutting of
and engraving of channels on PMMA, and thermal
and UV bonding for sticking the two layers.
Apart from developing a PMMA microchip, we
also developed a simple setup for fluorescence
detection. Majorly, the strategy employed for
detection of various analytes using aptamers
involves simple hybridization with dye labeled
complementary nucleic acid or sandwich assay.
Here, we have used both the strategies to check our
fluorescence detection setup. Aptamers were
immobilized using hexamethylene diamine (HMDA)
and glutaraldehyde for both the strategies. For
hybridization, we used an aminated sequence of
aptamer whose complementary 6-carboxyfluorescein
(FAM) labeled strand was made to interact. While in
other strategy, thrombin was used as a model analyte
to validate sandwich assay using our setup. The
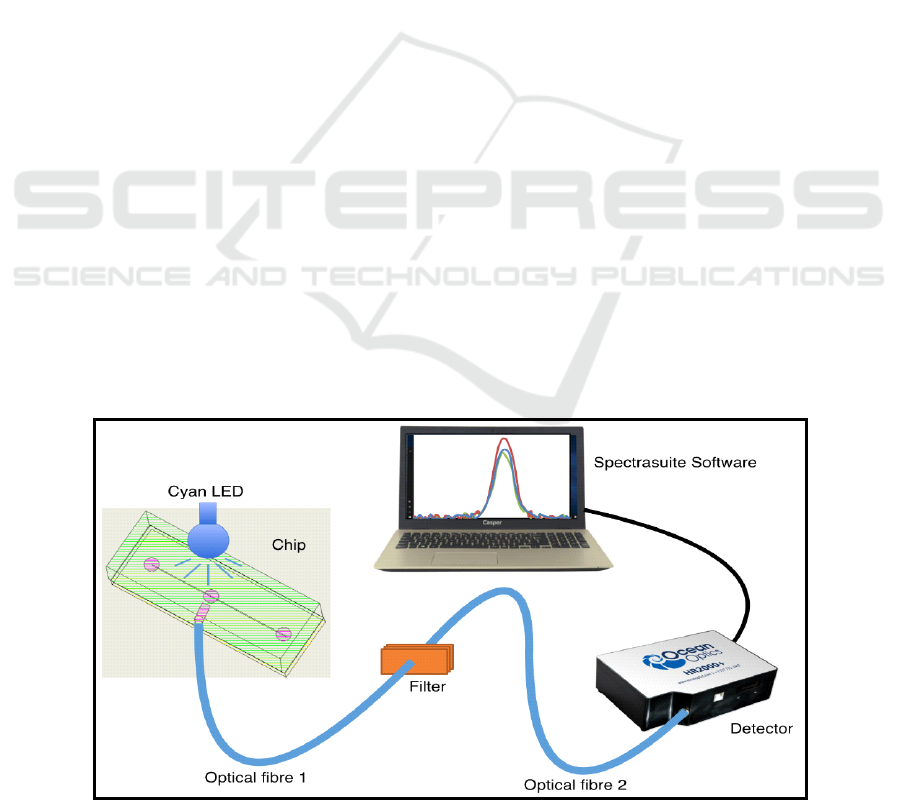
setup for fluorescence detection is shown in Figure
1. This setup enabled low cost fluorescence
detection without use of bulky microscopes. In
addition, this can be miniaturized further via using a
different detector and LabVIEW software.
2 MATERIALS AND METHODS
2.1 Materials
All chemicals were of analytical grade and
purchased from Sigma or Merck. Aptamer sequences
were purchased from Integrated DNA Technologies
(IDT). Sequences for first and second strategy were :
5’-/5AmMC6/GCC AAA TTG TTT GAC GAG A-
3’, 5’-/56-FAM/ TCT CGT CAA ACA ATT TGG
C-3’ and 5’-/5AmMC6/ TTT TTG GTT GGT GTG
GTT GG-3’, 5’-/56-FAM/TTT TTT TTT TTT TTT
AGT CCG TGG TAG GGC AGG TGG GGG TGA
CT-3’. Cyan LED 490nm, PMMA sheets and optical
narrow bandpass interference filter of 530 nm were
purchased from eBay India Online Store and Optics
& Allied Engg. Pvt. Ltd. respectively.
2.2 PMMA-PMMA Chip Fabrication
PMMA sheets of thickness 1.5 and 5 mm were cut
into substrates of 60 x 20 mm using CO
2
laser
cutting machine. Same technique was used to
engrave channel of depth 130µm, puncture for inlet
tubing, sensing area and outlet tubing, and detection
area. The detection area was made in such a way that
it will be at 90º angle with LED source for
fluorescence detection. Further, substrates were
ultrasonicated in Isopropyl alcohol (IPA) for 20
minutes and dried using nitrogen purging.
For bonding of both layer of substrates, UV and
Figure 1: Fluorescence detection setup used with microfluidic chip.
Microfluidic Platform for Aptamer based Fluorimetric Analysis of Analytes
219
solvent assisted thermal bonding were used. First,
ethanol was poured on both substrates, aligned over
each other avoiding any air bubble, clipped properly
and kept for UV bonding for 20 minutes. Then, after
removing clips, chips were kept on hot plate for 2
hours under 1 Kg weight at 120ºC. Further, weights
were removed when substrate cooled to room
temperature. Next, they were soaked in IPA for 10
minutes and rinsed with double distilled water.
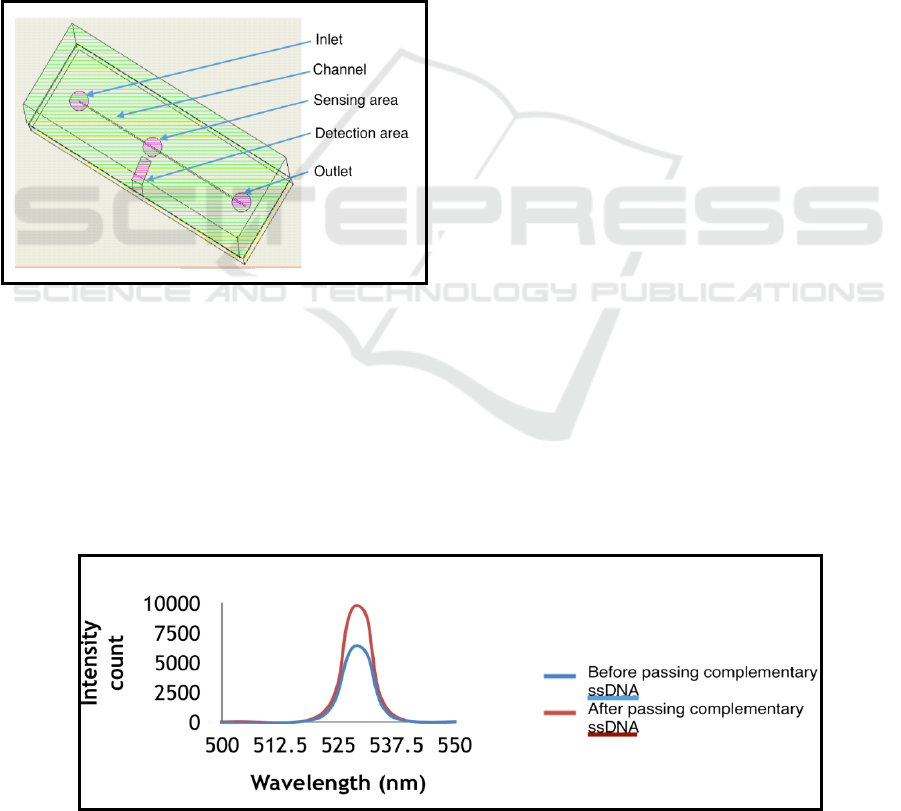
Design of PMMA-PMMA microfluidic chip is
shown in Figure 2. Here, detection area was an
aperture in PMMA substrate at 90º to sensing area
where optical fiber was inserted for fluorescence
detection. Sensing area is the area where aptamers
were immobilized for interaction with analyte. Inlet
and outlet in the microchip were made for insertion
of inlet and waste tubing.
Figure 2: Design of PMMA-PMMA microfluidic chip.
2.3 Sensing Area Modification and
Aptamer Immobilization
Sensing area on PMMA was treated with a solution
of 10% HMDA in 100 mM borate buffer of pH 11.5
for 2 hours. Following this, the surface of sensing
area was washed 3-4 times with distilled water.
Afterwards, surface was activated with 2.5%
glutaraldehyde in 0.1 M phosphate buffer pH 7 for 2
hours at room temperature followed by washing with
0.1 M phosphate buffer. Then, aminated aptamer
sequences were drop casted on sensing area for
immobilization and kept for 1 hour at room
temperature. Next, channels were washed with 1×
Tris EDTA (TE) buffer using syringe pump with
flow rate of 5 µl/min. In our two different strategies,
different sequences were immobilized using same
protocol. Immobilization step was confirmed by
immobilizing an aptamer 5’-/5AmMC6/GCC AAA
TTG TTT GAC GAG A-3’ and passing FAM
labeled complementary strand 5’-/56-FAM/ TCT
CGT CAA ACA ATT TGG C-3’. Fluorescence
intensity from sensing zone was checked before and
after passing complementary strand into the
microchannel.
2.4 Instrumentation
The instrumentation involved in our setup had a
Cyan LED of 490nm as excitation source. LED was
placed just above sensing area which was at 90º to
detection area. Optical fiber was inserted into
detection area of PMMA-PMMA chip followed by
an optical narrow bandpass interference filter of 530
nm. This optical fiber passed fluorescence signal
picked from the sensing zone of the microchip. Next,
the signal from filter was further collected at the
photodetector (Ocean Optics HR2000+) and changes
were read via commercial Spectrasuite software
from Ocean optics.
2.5 Sensing Procedure
In our first strategy, immobilized aptamer 5’-
/5AmMC6/GCC AAA TTG TTT GAC GAG A-3’
was made to interact with different concentrations of
complementary FAM labeled DNA 5’-/56-FAM/
TCT CGT CAA ACA ATT TGG C-3’. Concentra-
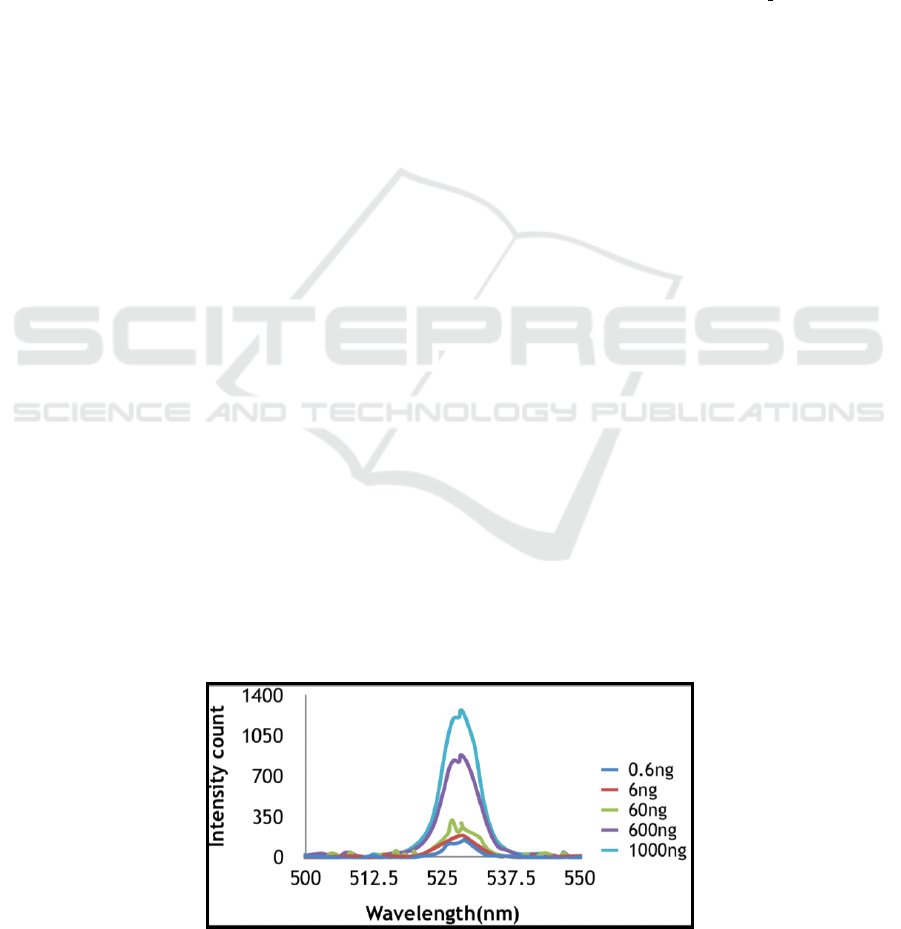
Figure 3: Increase in fluorescence after passing FAM labeled complementary ssDNA.
BIODEVICES 2018 - 11th International Conference on Biomedical Electronics and Devices
220
tion from 0.6-1000ng of complementary DNA was
passed through channel for 45 minutes and then
channels were washed with 1×TE buffer for 15
minutes. Both steps were performed at 5µl/min.
Fluorescence was noticed before and after passing
complementary DNA.
For confirmation of sandwich assay protocol,
immobilized primary aptamer 5’-/5AmMC6/ TTT
TTG GTT GGT GTG GTT GG-3’ was made to
interact with model analyte thrombin. Different
concentrations of thrombin, from 6-600 ng were
passed through channel for 45 minutes and then
channels were washed using 1×TE buffer for 15
minutes. Both steps were performed at 5 µl/min.
Afterwards, FAM labeled secondary aptamers 5’-
/56-FAM/TTT TTT TTT TTT TTT AGT CCG TGG
TAG GGC AGG TGG GGG TGA CT-3’ were
passed over the detection zone. Fluorescence
intensities from detection zone were recorded before
and after passing secondary aptamers.
3 RESULTS AND DISCUSSION
3.1 Confirmation of Sensing Protocol
We checked immobilization of aptamer by passing
its FAM labeled complementary strand in the
microchannel post immobilization of aptamer in the
sensing area. We found an increase in fluorescence
intensity post hybridization as shown in Figure 3.
This increase shows that hybridization occurred on
binding of FAM labeled complementary DNA strand
which increased number of FAM molecules in
sensing area. Hence, it proved successful
immobilization of aptamer.
Further, we tested our first strategy of
fluorescence detection using simple hybridization of
FAM labeled complementary strand. Increase in
fluorescence was seen with increase in concentration
of complementary ssDNA as shown in Figure 4. A
linear calibration curve was also obtained for the
same, as shown in Figure 5. Both figures show
proportionate increase in fluorescence with increase
in concentration of complementary DNA strand.
When complementary DNA strand was passed
through channel of microfluidic chip, it hybridized
with immobilized aptamer. According to the
arrangement in our setup, when FAM labeled
complementary DNA was targeted with light of cyan
LED source, dye molecules got excited and showed
fluorescence which was detected at 90º angle to
detection area using attached fiber optic probe and
coupled CCD array spectrometer. Therefore, it
proved direct relationship between concentration of
analyte and fluorescence. According to figure 5, the
linear range of detection was between 6 to 1000 ng
of ssDNA with 6 ng as the practical LOD for
measurement. Hence, this setup could be used for
detection of nucleic acid sequences using simple
hybridization.
3.2 Sandwich Assay Protocol
As our second detection strategy, we tested
fluorescence detection using thrombin protein to
validate sandwich assay using primary and FAM
labeled secondary aptamers. Again, increase in
fluorescence was observed proportionate to the
concentration of analyte thrombin as seen in Figure
6. When thrombin was passed through microchannel
in microfluidic chip, it interacted with its
immobilized primary aptamer. Then, FAM labeled
secondary aptamer for thrombin was passed through
channel. This secondary aptamer attached wherever
thrombin analyte was bound to primary aptamer and
hence, any increase in fluorescence was directly
related to thrombin concentration, as secondary
aptamer would not bind in absence of thrombin in
sensing area. As per figure 6, the linear range of
detection was found as 125-600 ng with 125ng as the
practical LOD for measurement. Therefore, the
Figure 4: Increase in fluorescence proportionate with concentration of complementary ssDNA.
Microfluidic Platform for Aptamer based Fluorimetric Analysis of Analytes
221
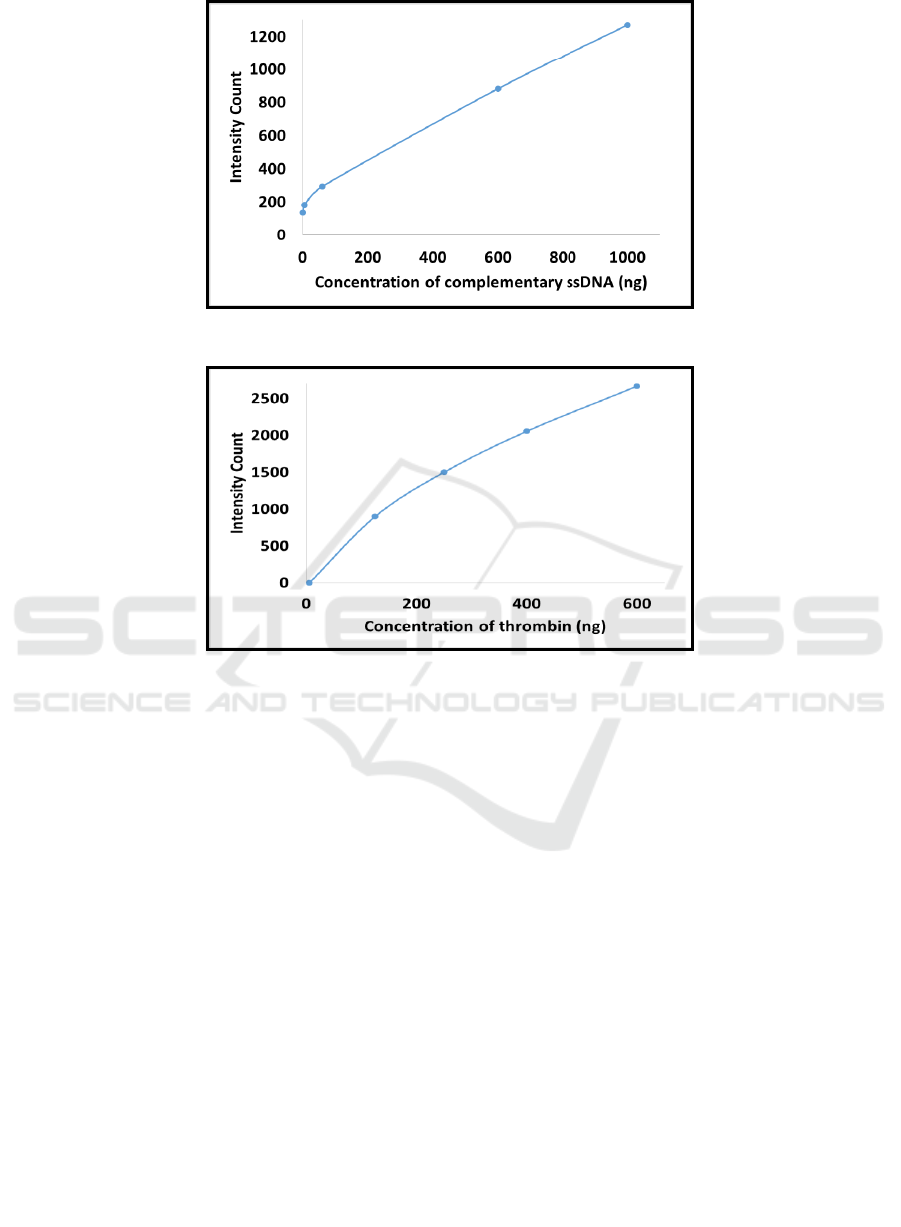
Figure 5: Calibration curve for complementary ssDNA.
Figure 6: Increase in fluorescence proportionate to increasing concentration of thrombin.
protocol was suitable for detection of diseases such
as some types of cancer, microbial diseases,
myocardial infarction etc. that gives rise to altered
concentration of biomarkers such as protein in blood
(Viscidi, 1987).
4 CONCLUSION
We have reported a simple and low cost setup for
fluorescence detection using aptamers as probe
without using any bulky devices like microscope.
We fabricated a PMMA-PMMA microfluidic chip
using simple and easily available laboratory
techniques and combined the chip with a simple
fluorescence detection setup. In this new approach,
we used two different strategies to use aptamers as
probe. In first strategy, diseases which can be
identified by nucleic acid hybridization could be
analyzed. We proved this using known sequence of
aptamer and its FAM complementary strand. Here,
we found that whole setup showed reliable linear
range of detection. In other strategy, diseases that
can be identified using analysis of any proteinic
biomarker could be detected using our sandwich
technique. In this, we used thrombin as model target
to validate our setup. We found that the minimum
concentration of thrombin which could be detected
out using our setup was impressive. Both the
strategies proved successful with our setup. In
future, this setup size could further miniaturized by
using a small CCD array, microcontroller and
LabVIEW software.
REFERENCES
Abu-Salah, K. M., Zourob, M. M., Mouffouk, F.,
Alrokayan, S. A., Alaamery, M. A., & Ansari, A. A.
(2015). DNA-based nanobiosensors as an emerging
platform for detection of disease. Sensors, 15(6),
14539-14568.
Fenzl, C., Hirsch, T., & Baeumner, A. J. (2016).
Nanomaterials as versatile tools for signal
amplification in (bio) analytical applications. TrAC
Trends in Analytical Chemistry, 79, 306-316.
BIODEVICES 2018 - 11th International Conference on Biomedical Electronics and Devices
222

Fixe, F., Dufva, M., Telleman, P., & Christensen, C. B. V.
(2004). Functionalization of poly (methyl
methacrylate) (PMMA) as a substrate for DNA
microarrays. Nucleic acids research, 32(1), e9-e9.
Song, K. M., Lee, S., & Ban, C. (2012). Aptamers and
their biological applications. Sensors, 12(1), 612-631.
Su, W., Gao, X., Jiang, L., & Qin, J. (2015). Microfluidic
platform towards point-of-care diagnostics in
infectious diseases. Journal of Chromatography
A, 1377, 13-26.
Tennico, Y. H., Hutanu, D., Koesdjojo, M. T., Bartel, C.
M., & Remcho, V. T. (2010). On-chip aptamer-based
sandwich assay for thrombin detection employing
magnetic beads and quantum dots. Analytical
chemistry, 82(13), 5591-5597.
Viscidi, R. P., & Yolken, R. G. (1987). Molecular
diagnosis of infectious diseases by nucleic acid
hybridization. Molecular and cellular probes, 1(1), 3-
14.
Wang, L., & Li, P. C. (2011). Microfluidic DNA
microarray analysis: a review. Analytica chimica acta,
687(1), 12-27.
Xu, Y., Yang, X., & Wang, E. (2010). Aptamers in
microfluidic chips. Analytica chimica acta, 683(1), 12-
20.
Microfluidic Platform for Aptamer based Fluorimetric Analysis of Analytes
223
