
A Risk-aware Access Control Model for Biomedical Research
Platforms
Radja Badji
1
and Fida K. Dankar
2
1
Independent Scientist, Lille, France
2
College of Information Technology, UAEU, AlAin, U.A.E.
Keywords: Privacy Preserving Data Sharing, Privacy Risk, Access Control Models.
Abstract: Data sharing and collaboration are important success factors for modern biomedical research. As biomedical
data contains sensitive information, any mechanism that governs biomedical data sharing should protect
subjects’ privacy while providing high-utility data in an efficient and prompt manner. The use of biomedical
data for research has been studied extensively from the legal aspect. Several regulations control its use and
sharing to limit privacy risks. However, current sharing mechanisms can be a barrier to the research
community needs. Going through the IRB process is time consuming and will become a bottleneck for the
intensive data need of the biomedical research community. Alternatively, creating a universal de-identified
research sub-dataset accessible through honest-broker-systems will not satisfy all research use-cases, as
stringent de-identification methods can reduce data utility. A risk-aware access control model is a good
alternative toward making data more available. In such a model, data requests are evaluated against their
incurred privacy risks, and are granted access after the application of appropriate protection levels. In this
paper, we describe a formal risk-aware model that will be used in the access control layer and describe the
different risk components that can be combined to provide a decision against a data access request.
1 INTRODUCTION
Data-sharing and collaboration are important key
success factors for modern biomedical research
(Lynch, 2011). The scientific community realized the
importance of data sharing and many international
initiatives are focusing on new policies and
procedures to promote biomedical data sharing
among the research community, such as, the
International Cancer Genome Consortium (ICGC)
(“International Cancer Genome Consortium,” n.d.)
and the Global Alliance for Genomics and Health
(GA4GH) (“Home | Global Alliance for Genomics
and Health,” n.d.). These initiatives reflect, on one
hand, the desire to capture a wide spectrum of
detailed biomedical data and on the other hand, the
need of collaborating across disciplines and
institutions.
This emerging context poses new challenges
toward safeguarding the privacy and security of the
data subjects, complicating the traditional institution-
based oversight system. Institutional Review Boards
(IRBs) or ethical committees are in charge of
analysing and evaluating research proposals in terms
of their risk and benefit. In general, the approved
research projects are the ones that present consequent
benefit with minimal risk and that comply with the
ethical standards and the local policies.
However, going through the IRB process is time
consuming and with the intensive data consumption
need of modern biomedical research, it is expected
that the IRB process will become a bottleneck for the
research activity (He et al., 2014). A current
mechanism to overcome the burden of the IRB
process is to share a universal de-identified sub-
dataset with the research community. An IRB-
approved honest broker system is in charge of the
distribution and the access to the de-identified data.
Although this mechanism reduces the time to acquire
data, it treats all requests equally in terms of de-
identification, thus disregarding the individualities of
the different data requests.
In a more efficient data-sharing scenario, the level
of granted access has to reflect the risk that we incur
with this data sharing and has to be compatible with
the research purpose. Moreover, it is not reasonable
to seek IRB approval for each research proposal
especially when it is in the exploratory phase. In
322
Badji, R. and Dankar, F.
A Risk-aware Access Control Model for Biomedical Research Platforms.
DOI: 10.5220/0006608403220328
In Proceedings of the 4th International Conference on Information Systems Security and Privacy (ICISSP 2018), pages 322-328
ISBN: 978-989-758-282-0
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

(Dankar and Al-Ali, 2015; Dankar and Badji, 2017),
the authors describe a theoretical multi-level privacy
protection framework for biomedical data
warehouses. The objective of the proposed
framework is to create a responsible data-sharing
mechanism through an electronic Honest Broker
System (e-HBS) that will enable the researchers’
timely access to biomedical data while lowering the
data access-related risk to an acceptable level.
Briefly, the cited model evaluates the risk posed by a
data request using all contextual information
surrounding the request and presents it to an access
control module that applies mitigation measures to
counter the posed risk.
Thus the risk-aware access control system
(RAAC) allows the mitigation of risks while previous
existing systems concentrate on the worst-case
scenario in the system design, and their access
decisions are consequently exclusively binary: allow
or deny. With the RAAC model, low-risk access
requests will be granted and high risk access requests
will be denied or granted after the enforcement of
some risk mitigation measures, such as: scaling-up
the security applying data de-identification, and/or
enforcing constraints on data access.
In this paper, we propose a formal risk aware
access-control model for the risk aware information
disclosure framework proposed in (Dankar and Badji,
2017). The model incorporates two use cases: (i) the
e-HBS case (described earlier), in which access
control decisions are made to counter the risk
associated with the different requests, and (ii) the IRB
case, which is the usual IRB manual application
process. The IRB use-case is useful for investigators
who are not happy with the level of protection offered
by the automated process, or the investigators who are
willing to endure the extra time taken by the IRB.
Our work extends prior work (Armando et al.,
2015; Chen et al., 2012) in risk aware access control
to the field of biomedical research. One of the
distinctive features in our model is the introduction of
the legal risk, which represents the decision of the
IRB. Specifically, our system allows the IRB as an
approved authority to override access decisions made
by the system at any point in time under specific
conditions. Thus realizing the above two scenarios.
The rest of the paper is organized as follows:
Section 2 describes a high-level overview of our
RAAC-based model; Section 3 presents the model
formally and defines the proposed authorization
function; Section 4 summarizes related work in the
field and compares it to our model; and Section 5
concludes the paper and presents future research
directions.
2 RISK-AWARE SYSTEMS FOR
BIOMEDICAL DATA SHARING
Privacy preservation and legislation compliance are
key aspects in the development of biomedical
research platforms. Research Data requests will be
granted, if and only if, the risk incurred by such access
is acceptable and controlled. In (Cheng et al., 2007)
the authors define the risk of a data request as a
predictive function of the expected value of damage
(equation 1).
Quantified risk = (probability of damage)×
(value of damage)
(1)
Risk estimation is of the responsibility of the
policy writer and is domain and application
dependent. In our case, the IRB is responsible of the
estimation. Nevertheless, our model has to be able to
define all the necessary components used in this
estimation.
In equation 1, the risk is associated with a
probability and represented as a metric value. The
damage in the equation can be caused by a number of
circumstantial factors that are specific to the data
sharing episode. In (Dankar and Badji, 2017), the
authors define and measure these factors, they
establish that the risk of granting data access to a user
is dependent upon the data requested, the stated
purpose for the access, the motives of the user (can
we trust the user?) and on the security of the user’s
environment (check Figure 1). For example, access to
highly sensitive data at the data-holder’s location by
a trusted user is inherently less risky than providing
the same user with a copy of the dataset. Similarly,
access to de-identified clinical data from a secure
remote system is inherently less risky than access to
identifiable data from an unknown location.
The authors then define the risk-aware access
control as realized on a risk scale divided into
multiple risk bands (or classes). These risk bands are
determined according to the organization risk
tolerance levels and can be dynamically adjusted.
Every access request falls into one of these risk bands
according to its risk estimate. The access decision is
made accordingly and will deny, allow with or
without risk mitigation-measures the access (See
Figure 1, which is an adaptation from (Cheng et al.,
2007)). For a detailed description of the different risk
dimensions, ways to calculate the risk and an
illustration, refer to (Dankar and Badji, 2017).
A Risk-aware Access Control Model for Biomedical Research Platforms
323
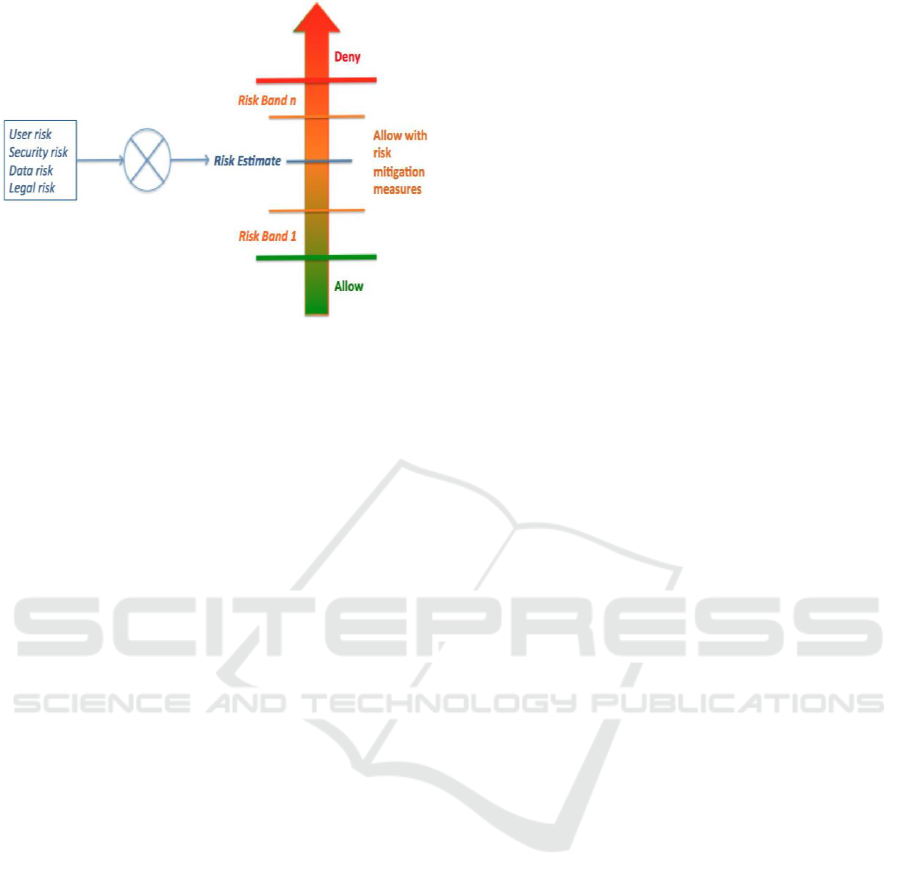
Figure 1: Risk scale associated with an access request.
3 RISK-AWARE ADAPTIVE
ACCESS CONTROL MODEL
We take the definition of the formal model for risk-
aware access control (RAAC) developed in (Chen et
al., 2012) and its subsequent refinement proposed in
(Armando et al., 2015) as the basis of our model. We
add a new function called IRBapproved to represent
the legal risk. This function allows the access in the
context of risky situations as long as, the IRB, as a
legal entity, has authorized that. We then define the
final data sharing decision as a combination of
individual risk functions. The components of the
formal model are defined as follows:
• U: a set of users.
• : denotes a dataset and denotes any
subset
• : is a set of operations that can be applied to a
dataset , denotes a particular operation.
• : a set of access purposes representing all the
possible access purposes in our platform and
is a particular data access purpose.
• : a set of permissions represented as operation-
data-purpose tuples .
• : a set of access requests represented as a pair
• : a set of risk mitigation methods. A risk
mitigation method is any action that can
be taken by the user or/and the system to reduce
risk. Such as data de-identification or constraints
on data access time and mode (Dankar and Badji,
2017).
• : a risk domain where 0
represents no risk and 1 is the maximum risk. A
risk interval
is defined as
•
: is a list that
associates risk levels with mitigation methods
and represents the risk mitigation strategy. Note
that a risk level can be associated with multiple
mitigation measures. The choice of what
mitigation measure to apply is left to the user.
• : a set of states represented as tuples of the form
where a state
represents a request in an
environment adopting strategy and
implementing rules . In our case, represents
the features of a Role-Based Access Control
model (RBAC) (Chen and Crampton, 2011). It
defines a set of roles , the user-role assignment
relation , the role permission
assignments relation , and the role
hierarchy . In other words, defines
who has the right to request data, and the kind of
data they can request based on their role.
•
: a boolean function that holds if
and only if is granted access to according to
.
• : an authorization function
where and for each
tuple , the IRB approval status
is returned. will be set
by default to 0, unless the user has an explicit
IRB approval for the data. This function reflects
somehow the legal risk of a granted data access.
If the IRB approves the request, then the legal
risk is minimal.
• : a risk function with
that returns for each access request in a
particular state , the risk
. This
function is the core of our system and will be
defined next.
• : an authorization decision function where
and for each access
request in a particular current state an
authorization decision
is
returned along with a set of mitigation methods
. Formally:
Several parameters can be taken into account in
the estimation of the risk associated with a data
ICISSP 2018 - 4th International Conference on Information Systems Security and Privacy
324

request (Chen and Crampton, 2011). In prior work
(Dankar and Badji, 2017), we identified the different
risk categories in the context of biomedical research
and data sharing. We proposed also different methods
to calculate the different risks associated to these
categories. The aim of this model is to use the values
of these different risks to calculate an access decision.
We define briefly each risk category and then we
propose an equation () to combine them:
− User risk: the user risk is related to the
trustworthiness of the user. The risk associated to
the user trustworthiness is defined as follows:
where
reflects the degree of trustworthiness
of user .
− Security risk: is the risk associated with the
request session, such as: network security,
connection location, connection time, etc.
Defining a precise method to calculate this kind
of risk is not an easy task. Nevertheless, several
works in the literature have proposed methods to
calculate such risk (“Google Android: A
Comprehensive Security Assessment - Google
Scholar,” n.d.). It is out of the scope of this paper
to adopt one of these methods. However, we use
the formal definitions of the associated risks to
define our model.
where reflects the security level of the
request session under the current state .
− Data sensitivity risk: There are many ways to
estimate the data sensitivity: data classification,
statistical metrics, etc [8]. The formal definition
of the risk associated to data sensitivity is as
follows:
Where reflects the data sensitivity level.
− Access purpose risk: we consider that in
biomedical research some tasks are riskier than
others. For example, exploratory research is less
risky than the creation of public data sets out of
the requested data. Whenever the data request
could be followed by data publication the risk has
to be higher. This necessitates of course a
classification of the different data access
purposes and the estimation of their incurred risk.
Hereafter is a formal definition of the access
purpose risk that will be used in our model:
Where reflected the degree of risk
associated with the access purpose .
− Legal risk: is reflected with the function
.
The IRB is the authority that will define how we
calculate the risk associated to each situation and the
combination of them. The combined risk will be
defined as a function of the different risk elements.
The authorization decision is made based on the
combined risk estimation:
(2)
Where
Using Equation 3, we take the approach of
maximizing the risk. If the data is very sensitive and
the researcher has the IRB approval, she will be able
to access the data. By the same manner, we can link
the other risk components with the IRB approval. For
example, a less trusted person will be able to access
sensitive data if she has the IRB approval and so on.
Embedding the IRB approval into the equation will
allow us to check for the IRB approval for every
access and the privileges will be revoked as soon as
the IRB approval is suspended or expired for
example. The IRB approval will be able to lower the
risk associated to the data access purpose, the data
sensitivity and the user trustworthiness but not the
security one. Even if a researcher has an IRB approval
we cannot take the risk of disclosing sensitive data
into an unsecure connection for example.
A Risk-aware Access Control Model for Biomedical Research Platforms
325

3.1 Illustration
Assume that a biomedical platform holds different
data sets of different sensitivity levels ranging fro 0
to 1 (with 1 being the highest sensitivity), such as:
−
: a public data set with sensitivity level 0
−
: a de-identified dataset of flu patients with
low sensitivity level 0.2
−
: a de-identified dataset of kids with ADHD
with sensitivity level 0.5
−
: a de-identified dataset of HIV patients with
high sensitivity level 0.8
When a researcher requests a permission to access
a dataset, Equation 3 is used to calculate the risk
associated with her request. The decision is made
according to the risk band corresponding to the
request risk level. For this example, let us suppose
that the researcher is highly trusted and is requesting
data for exploratory research purposes. Therefore, the
risks associated with the user trustworthiness and the
access purpose are minimal (say both are equal to 0).
Assume that the researcher is trying to access dataset
which is highly sensitive.
If the role associated to the researcher doesn’t
allow her to access the data set and she has no IRB
approval to do so, then the risk to access this data is
maximum = 1 and therefore has to be denied.
Otherwise, a risk calculation will be performed
according to the equation 3:
We have two possible cases:
− The researcher has IRB approval for this
particular dataset. Therefore:
, and
It means that the only risk considered here is
the security risk to avoid the disclosure of
sensitive data in case of high security risk.
− The researcher does not have IRB approval for
this particular data set. Therefore:
, and:
In this situation, we choose the maximum of the
risks related to data sensitivity and security.
4 RELATED WORK
With the pressing need of data sharing and
collaborative data computing there is a growing
interest from the scientific community to develop
novel data-access models and efficient data sharing
mechanisms while maintaining data privacy and
legislation compliance. In the literature, the notion of
risk in the context of access control is of two types:
1. First, the risk of hindering the performance of a
task if the information is not disclosed. For
example, the exceptional disclosure of sensitive
medical information in some urgent situation
like in (Choi et al., 2015) or (Kayes et al.,
2015).
2. Second, the risk of privacy leakage associated
with data disclosure.
In our case, we focus on the second point, that is,
the risk associated with data disclosure.
In existing systems, the risk is either explicitly
taken into account by the system by having a risk
calculation function or implicitly taken into account
by enforcing context-aware policies, where the
context captures some risk-related situation
information. Our proposed model is based on the
formal meta-model developed in (Chen et al., 2012)
The instantiation of the meta-model allows the
creation of RAAC models with different risk
mitigation strategies. The authors focus on system
obligations and user obligations as risk mitigation
strategies and propose the models to implement these
strategies.
In (Armando et al., 2015), the authors consider
the risk of leaking privacy-critical information when
querying a dataset. The risk is calculated according to
the query result and using anonymity metrics related
to personal-identifiable information. The model
includes adaptive anonymization operations as risk
mitigation methods to lower the risk associated with
a particular data request. In (Kandala et al., 2011), the
authors rely on an attribute-based framework to
capture the different elements needed to implement
an adaptive risk-based access control mechanism.
The attributes capture information about different
components that can impact the risk level associated
with a particular data request. These components are
related to the data access purpose, the security level
and the situational factors reflecting any contextual
factors that can increase the risk related to a data
request.
ICISSP 2018 - 4th International Conference on Information Systems Security and Privacy
326

5 CONCLUSIONS/FUTURE
WORK
In this paper we depicted the need for risk-aware
access control models that support the regulation,
development, and deployment of access control
procedures for data sharing in biomedical research
platforms. We proposed a method that identifies the
essential risk components, necessary for such access
control procedures and extended existing models to
overcome the limitations of the “manual” biomedical
data sharing processes, such as the IRB, and the
“automated” ones based on e-HBS.
Currently we are working on coming up with
efficient equations to calculate the different risk
elements. This work is challenging and requires
significant efforts on many fronts:
• Assigning data sensitivity to datasets is the main
challenge. As a start, we are currently working
on classifying data into a set of pre-defined
sensitivity classes.
• Creating local (and ideally universal) user
records for storing data breach information is
another theoretical/practical challenge.
Analogous to credit scores, the risk associated
with individual users should indicate the gravity
of their past breaches, and should reward users’
progress. Our approach is to standardize all data
breaches (i.e. create a breach classification) and
create an account system for all users that can be
accessed by data holders when required.
• The security of the user’s environment is related
to the user’s institution (the research institution
to which a user is affiliated). Thus, the risk can
benefit from having universal security
certification programs for research institutions.
Such programs would provide certifications to
different institutions based on their privacy and
security practices. Refer to (El Emam et al.,
2009) for a list of parameters to take in
consideration when evaluating institutions’
privacy and security practices.
Another necessary task is to extend the system to
provide Omics data. For that, we need to study the re-
identification power of this data to be able to annotate
it with any privacy risk. Some work has already been
done along these lines for single nucleotide
polymorphisms (SNPs) (Lin et al., 2004).
REFERENCES
Armando, A., Bezzi, M., Metoui, N., Sabetta, A., 2015.
Risk-Aware Information Disclosure, in: Garcia-Alfaro,
J., Herrera-Joancomartí, J., Lupu, E., Posegga, J.,
Aldini, A., Martinelli, F., Suri, N. (Eds.), Data Privacy
Management, Autonomous Spontaneous Security, and
Security Assurance, Lecture Notes in Computer
Science. Springer International Publishing, pp. 266–
276.
Chen, L., Crampton, J., 2011. Risk-aware role-based access
control, in: International Workshop on Security and
Trust Management. Springer, pp. 140–156.
Chen, L., Crampton, J., Kollingbaum, M. J., Norman, T. J.,
2012. Obligations in risk-aware access control, in:
Privacy, Security and Trust (PST), 2012 Tenth Annual
International Conference on. IEEE, pp. 145–152.
Cheng, P.-C., Rohatgi, P., Keser, C., Karger, P.A., Wagner,
G.M., Reninger, A.S., 2007. Fuzzy multi-level security:
An experiment on quantified risk-adaptive access
control, in: 2007 IEEE Symposium on Security and
Privacy (SP’07). IEEE, pp. 222–230.
Choi, D., Kim, D., Park, S., 2015. A framework for context
sensitive risk-based access control in: medical
information systems. Comput. Math. Methods Med.
2015.
Dankar, F. K., Al-Ali, R., 2015. A Theoretical Multi-level
Privacy Protection Framework for Biomedical Data
Warehouses. Procedia Comput. Sci., The 6th Interna-
tional Conference on Emerging Ubiquitous Systems
and Pervasive Networks (EUSPN 2015)/ The 5th
International Conference on Current and Future
Trends of Information and Communication Technolo-
gies in Healthcare (ICTH-2015)/ Affiliated Workshops
63, 569–574. doi:10.1016/j.procs.2015.08.386
Dankar, F. K., Badji, R., 2017. A risk-based framework for
biomedical data sharing. J. Biomed. Inform. 66, 231–
240.
El Emam, K., Dankar, F. K., Vaillancourt, R., Roffey, T.,
Lysyk, M., 2009. Evaluating the risk of re-identification
of patients from hospital prescription records. Can. J.
Hosp. Pharm. 62, 307.
Google Android: A Comprehensive Security Assessment -
Google Scholar [WWW Document], n.d. URL
https://scholar.google.ae/scholar?q=Google+Android
%3A+A+Comprehensive+Security+Assessment&btn
G=&hl=en&as_sdt=0%2C5 (accessed 5.21.17).
He, S., Narus, S. P., Facelli, J. C., Lau, L. M., Botkin, J. R.,
Hurdle, J. F., 2014. A domain analysis model for eIRB
systems: Addressing the weak link in clinical research
informatics. J. Biomed. Inform. 52, 121–129.
Home | Global Alliance for Genomics and Health [WWW
Document], n.d. URL http://genomicsandhealth.org/
(accessed 5.21.17).
International Cancer Genome Consortium [WWW
Document], n.d. URL http://icgc.org/ (accessed
5.21.17).
Kandala, S., Sandhu, R., Bhamidipati, V., 2011. An
attribute based framework for risk-adaptive access
control models, in: Availability, Reliability and Security
A Risk-aware Access Control Model for Biomedical Research Platforms
327

(ARES), 2011 Sixth International Conference on. IEEE,
pp. 236–241.
Kayes, A. S. M., Han, J., Colman, A., 2015. OntCAAC: an
ontology-based approach to context-aware access
control for software services. Comput. J. bxv034.
Lin, Z., Owen, A. B., Altman, R. B., 2004. Genomic
Research and Human Subject Privacy. Science 305,
183–183. https://doi.org/10.1126/science.1095019
Lynch, R. P., 2011. Collaborative Innovation: Essential
Foundation of Scientific Discovery, in: Ekins, S.,
Hupcey, ggie A. Z., Williams, A. J. (Eds.),
Collaborative Computational Technologies for
Biomedical Research. John Wiley & Sons, Inc., pp. 19–
37. doi:10.1002/9781118026038.ch2
ICISSP 2018 - 4th International Conference on Information Systems Security and Privacy
328
