
Real-time Low SNR Signal Processing for Nanoparticle Analysis with
Deep Neural Networks
Jan Eric Lenssen
1
, Anas Toma
2
, Albert Seebold
1
, Victoria Shpacovitch
3
, Pascal Libuschewski
1
,
Frank Weichert
1
, Jian-Jia Chen
2
and Roland Hergenr
¨
oder
3
1
CS VII - Computer Graphics, TU Dortmund University, Otto-Hahn-Straße 16, 44227 Dortmund, Germany
2
CS XII - Embedded Systems, TU Dortmund University, Otto-Hahn-Straße 16, 44227 Dortmund, Germany
3
Leibniz-Institute for Analytical Science, ISAS e.V., Bunsen-Kirchhoff-Straße 11, 44139 Dortmund, Germany
Keywords:
Nanoparticle Analysis, Deep Learning, Convolutional Neural Network, GPGPU Real Time Processing,
Biosensing.
Abstract:
In this work, we improve several steps of our PLASMON ASSISTED MICROSCOPY OF NANO-SIZED OBJECTS
(PAMONO) sensor data processing pipeline through application of deep neural networks. The PAMONO-
biosensor is a mobile nanoparticle sensor utilizing SURFACE PLASMON RESONANCE (SPR) imaging for
quantification and analysis of nanoparticles in liquid or air samples. Characteristics of PAMONO sensor
data are spatiotemporal blob-like structures with very low SIGNAL-TO-NOISE RATIO (SNR), which indicate
particle bindings and can be automatically analyzed with image processing methods. We propose and evaluate
deep neural network architectures for spatiotemporal detection, time-series analysis and classification. We
compare them to traditional methods like frequency domain or polygon shape features classified by a Random
Forest classifier. It is shown that the application of deep learning enables our data processing pipeline to
automatically detect and quantify 80 nm polystyrene particles and pushes the limits in blob detection with very
low SNRs below one. In addition, we present benchmarks and show that real-time processing is achievable on
consumer level desktop GRAPHICS PROCESSING UNITs (GPUs).
1 INTRODUCTION
The effect of SURFACE PLASMON RESONANCE
(SPR) is often utilized to study interactions between
different types of biomolecules (nucleic acids, pep-
tides, lipids, proteins, etc.) and to determine con-
centrations and affinity constants of biomolecules in
solutions. The high sensitivity of SPR has led to a
common use of SPR sensors for real-time measure-
ments of biomolecule binding efficiency. However,
the task to quantify individual biological nanoparti-
cles with SPR sensors remained unsolved for a long
time.
Recently, the PLASMON ASSISTED MI-
CROSCOPY OF NANO-SIZED OBJECTS (PAMONO)
sensor was shown to overcome the limitation of
SPR to quantify individual biological nanoparticles
(Zybin, 2010; Zybin, 2013; Shpacovitch et al.,
2015): single viruses, virus-like particles and other
nanoparticles can be detected in suspensions of liquid
or air.
Manually analyzing the sensor data and quantify
the particles is a time-consuming task. An evalua-
tion of a single data set by an expert with a few hun-
dred particles can take several hours. The application
of a highly optimized GENERAL-PURPOSE COMPUT-
ING ON GRAPHICS PROCESSING UNITS (GPGPU)
pipeline (Siedhoff, 2016; Libuschewski, 2017) makes
it possible to automatically analyze the sensor data
and quantifying the nanoparticles in less than three
minutes. This enables the real-time measurements
of SPR sensors also for the PAMONO sensor. For
this automatic analysis, it was shown that it can re-
liably detect signals with an SNR down to 1.2 and
therefore, virus-like and polystyrene particles down
to 100 nm in our experiment setup (Siedhoff, 2016;
Libuschewski, 2017; Siedhoff et al., 2014). Automat-
ically detecting SPR signals with lower SNR has yet
to be accomplished.
In this work, we push the limits of our meth-
ods and move towards the goal of detecting particle
signals with an SNR below one in PAMONO sen-
sor data by incorporating three different deep neural
networks for nanoparticle analysis into our GPGPU
36
Lenssen, J., Toma, A., Seebold, A., Shpacovitch, V., Libuschewski, P., Weichert, F., Chen, J-J. and Hergenröder, R.
Real-time Low SNR Signal Processing for Nanoparticle Analysis with Deep Neural Networks.
DOI: 10.5220/0006596400360047
In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2018) - Volume 4: BIOSIGNALS, pages 36-47
ISBN: 978-989-758-279-0
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

Figure 1: Principle of operation of the PAMONO sensor. It consists of a laser, the flow cell, gold plate, and prism, and the
camera, as it is shown in the scheme. Nanoparticles attached to the gold layer result in refractive index changes of the surface,
which can be observed by the camera. On the captured image sequences, temporal intensity steps appear on particle binding
spots. Figure adapted from (Libuschewski, 2017).
pipeline: a spatiotemporal FULLY CONVOLUTIONAL
NETWORK (FCN) (Long et al., 2015), a time-series
analysis network (both for detection) and a CON-
VOLUTIONAL NEURAL NETWORK (CNN) (LeCun
et al., 1995) for classification. In our experimental
setup, this enables the quantification of 80 nm parti-
cles.
We evaluate those methods by performing two dif-
ferent types of experiments: a standalone classifica-
tion evaluation where we test and compare classifica-
tion methods on a dedicated benchmark data set and
a sensor data experiment where we apply our whole
processing pipeline on real sensor data. We show that
the proposed methods are able to achieve reliable re-
sults for sensor data containing particle signals with
an SIGNAL-TO-NOISE RATIO (SNR) below one, and
that the pipeline still fulfills the soft real-time property
on current GRAPHICS PROCESSING UNITs (GPUs),
which means that on average, the data is processed
with at least the same speed as the sensor provides it.
2 PAMONO-Biosensor
The PAMONO sensor utilizes a Kretschmann’s
scheme (Kretschmann, 1971) of plasmon excitation to
detect individual particles. In Kretschmann’s config-
uration, as shown in Figure 1, an incident laser beam
passes through a glass prism, which is coated with
a very thin gold film on one side. This film forms
a sensor surface, on which the interactions between
biomolecules occur. At a certain angle of incidence
(resonance angle), an incidence beam is not reflected
and the gold sensor surface is very sensitive to any
changes of the refractive index near it. Any changes
of the refractive index in close vicinity of the gold
interface result in changes of reflection conditions.
Thus, any binding of a particle to the gold surface re-
stores the local reflection on the binding spot.
The data characteristic of particle signals in cap-
tured images, as shown in Figure 1, is as follows: On
places with plasmon excitation through particle bind-
ings, an increase of intensity in the time dimension
(intensity step) can be observed in the corresponding
pixels. In the spatial dimensions, these pixels form
a blob with surrounding wave-like structures. Both
variations in intensity are indications for a particle
binding and can be detected and analyzed by image
processing methods.
3 RELATED WORK
In the following we provide a broad context for
nanoparticle analysis in Section 3.1, before we out-
line the related work for our data processing methods.
Analyzing PAMONO sensor data is most related to
the field of low SNR blob detection. Therefore, we
give a short overview about this subject in Section 3.2.
3.1 Nanoparticle Detection
When comparing SPR-based approaches to study bio-
logical nanoparticles, one should highlight the follow-
ing differences. Conventional SPR sensors deal with
the formation of a layer of biomolecules or biopar-
ticles onto a gold sensor surface and harnesses the
integral changes of reflectivity conditions to char-
acterize the layer assembly process. In contrast,
the PAMONO sensor utilizes local changes of re-
flectivity to show individual biological nanoparti-
cles. Firstly, the latter issue makes the PAMONO
sensor more sensitive in the detection and quan-
tification of biological nanoparticles. Secondary,
the PAMONO sensor provides direct information
about particle binding events. This helps to ob-
viate complex calculations of particle concentra-
tions based on the thickness of the particle layer
Real-time Low SNR Signal Processing for Nanoparticle Analysis with Deep Neural Networks
37

formed onto the sensor surface. Examples of other
nanoparticle analysis methods are SURFACE PLAS-
MON RESONANCE IMAGING (SPRi) (Steiner and
Salzer, 2001), NANOPARTICLE TRACKING ANALY-
SIS (NTA) (Dragovic et al., 2011), plaque assay (Dul-
becco, 1954), and ENZYME-LINKED IMMUNO-SOR-
BENT ASSAY (ELISA) (Gan and Patel, 2013). A
comprehensive overview about the plasmon reso-
nance effect is given by Pattnaik (Pattnaik, 2005).
Most similar to the PAMONO sensor are SPRi
sensors which are a wide spread technology that are
applied in a large field of applications (Beusink et al.,
2008; Chinowsky et al., 2004; Giebel et al., 1999;
Naimushin et al., 2003; Scarano et al., 2011). Steiner
et al. states that the reason for the advantage of SPRi-
based methods is that they show specific bounds of
unlabeled molecules under in-situ conditions (Steiner
and Salzer, 2001), which also holds for the PAMONO
sensor.
3.2 Low SNR Blob Detection
Low SNR blob detection is the most related task to
our PAMONO data analysis, as small blob-like struc-
tures need to be identified in gray-scale images. Most
methods that are comparable to our pipeline can han-
dle an SNR down to four (Cheezum et al., 2001) or
two (Smal et al., 2009)
Automatic tumor detection in breast ultrasound
images represents a similar task to PAMONO image
processing. Moon et al. (Moon et al., 2013) compute
features by convolving partial derivatives of a Gaus-
sian distribution with the input images to solve this
task and Liu et al. (Liu et al., 2010) apply this method
to detect blobs in natural scenes.
Another related task is finding blobs in images
from fluorescence microscopy, which was surveyed
by Cheezum et al. (Cheezum et al., 2001). It should
be noted that for the analyzed tracking tasks, an SNR
of four was required for the surveyed algorithms to
succeed. For PAMONO signals however, SNRs be-
low one need to be handled.
For live-cell fluorescence microscopy, different
spot detection methods have been surveyed by Smal
et al. (Smal et al., 2009). It is stated that most classical
methods need an SNR of four to work. After present-
ing algorithms for SNRs down to two, they recom-
mend using supervised machine learning methods for
data with low SNR. The deep neural networks used in
this work fall under this category.
4 METHODS FOR AUTOMATED
NANOPARTICLE DETECTION
The following section details our methods for
nanoparticle analysis. First, an overview about the
data processing pipeline is given in Section 4.1. In
Section 4.2 we describe our deep neural network
models for detection and classification and in Sec-
tion 4.3 we outline our frequency domain methods for
comparison. Last, implementation details are given in
Section 4.4.
4.1 Image Processing Pipeline -
overview
An overview of the image processing pipeline is given
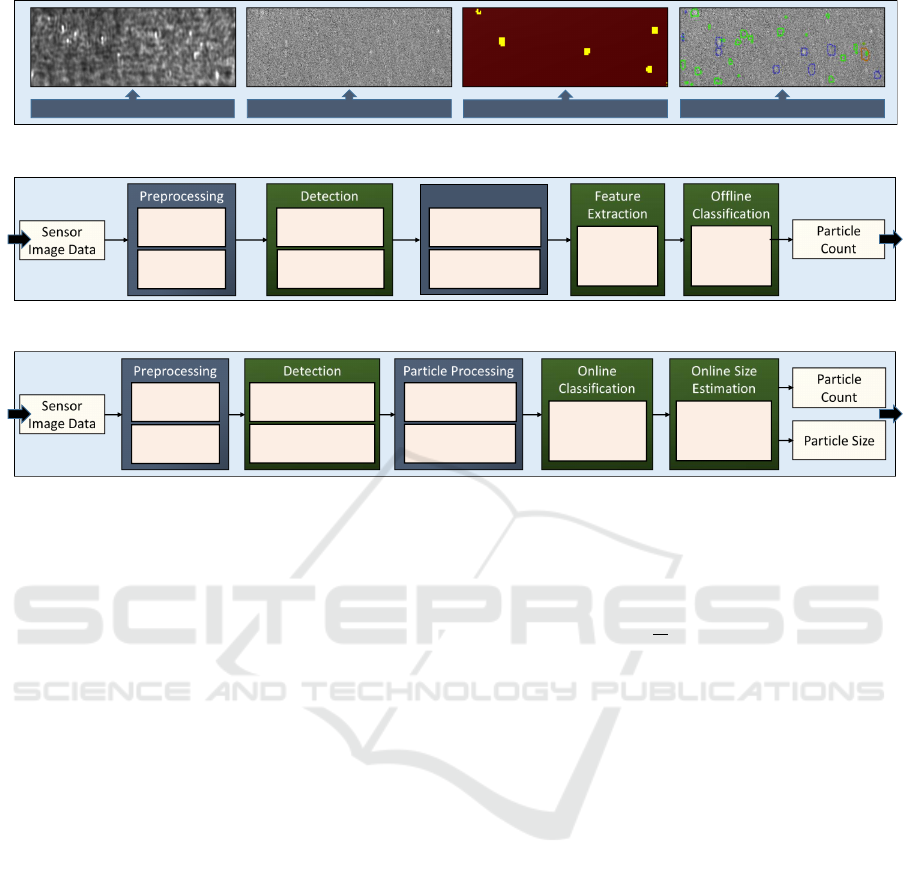
in Figure 2. Figure 2a shows the overall detection task
with example images: First, a raw image. Second,
an image with removed background. Third, detected
particle pixels as binary heat map. Finally, generated
particle candidates.
Figure 2b shows the existing PAMONO sensor
data processing pipeline from previous work (Sied-
hoff, 2016; Libuschewski, 2017), which makes use
of several traditional image processing methods. It
consists of the steps preprocessing, detection, particle
processing, feature extraction and offline classifica-
tion. The input is a sequence of sensor images and
the output is the particle count and the spatiotempo-
ral coordinates of each particle. The detection step
estimates a binary heat map marking possible par-
ticle signal positions. After the detection step, the
heat map is further processed to generate polygon
proposals. These polygons are matched over time to
obtain particle candidates that are visible over sev-
eral frames. Subsequently, polygon features are ex-
tracted and the particle candidates are either classified
as true particle or artifact/noise in an offline step, us-
ing a Random Forest classifier (Breiman, 2001). This
pipeline consists of a large number of image process-
ing methods that can be chosen and configured by pa-
rameter sets (Libuschewski, 2017). In addition, Sied-
hoff developed the SynOpSis approach to automati-
cally optimize parameter sets towards specific tasks,
using synthetic sensor data (Siedhoff, 2016).
Figure 2c shows our proposed processing pipeline,
which incorporates novel methods from the field of
deep learning: a FULLY CONVOLUTIONAL NET-
WORK (FCN) and a time-series analysis replace the
detection step, a CONVOLUTIONAL NEURAL NET-
WORK (CNN) online classification replaces the fea-
ture extraction and classification step and an addi-
tional CNN for online size estimation is added. The
online particle size estimation network is able to de-
BIOSIGNALS 2018 - 11th International Conference on Bio-inspired Systems and Signal Processing
38
Detected Particle Pixels
Background Removed
Raw Data
Particle Candidates
(a) Detection task overview.
Background
Elimination
Template
Matching
Particle Processing
Candidate
Generation
Patch Extraction
Random
Forrest
Classifier
Noise
Reduction
Time-Series
Analysis
Polygon
Shape
Features
(b) Baseline processing pipeline from previous work.
Background
Elimination
Fully Convolutional
Network
Candidate
Generation
Patch Extraction
Convolutional
Neural
Network
Noise
Reduction
Convolutional
Neural
Network
Time-Series
Analysis Network
(c) Proposed pipeline with deep neural networks.
Figure 2: An overview of the detection task, the baseline processing pipeline and the proposed processing pipeline. Traditional
image processing methods have been replaced by deep neural networks.
rive the size of individual particles, which is part of
previous work (Lenssen et al., 2017) but mentioned
here for the sake of completeness. All networks are
detailed in the following section.
Both pipelines follow the signal model
I(x, y,t) = B(x, y) · (T · A)(x, y,t) +N(x, y,t) (1)
where I is the sensor image sequence, T ·A represents
particle and artifact signals and N is additive noise
(Siedhoff, 2016). Given the sensor image sequence
I, we can approximate the particle and artifact signal
T · A by removing the constant-over-time background
signal B. This is done by dividing the current image in
the sequence by the mean of a set of previous frames.
Thus, only the non-constant parts, particle signals, ar-
tifacts and noise remains in the images. Then, the de-
tection and classification steps aim to distinguish the
particle signal P from artifact A and noise N. In the
following sections we provide details of our proposed
methods.
4.2 Deep Neural Networks
We present three different neural network architec-
tures for marking pixels that belong to a particle (de-
tection) and to sort out false detections (classifica-
tion). For the detection task, we present two different
approaches which we evaluate against each other. All
networks are trained using the cross entropy loss
L = −
1
N
N
∑
i=1
y
i
· log
ˆ
y
i
, (2)
where
ˆ
y
i
is the softmax output of the network and y
i
is a binary one-hot vector indicating the correct class.
For the detection FCN, we compute the pixel-wise
loss and average over the N pixels of all images in
one mini-batch while for the remaining networks, the
cross entropy is only averaged over all N examples in
one mini-batch.
While choosing the neural network architectures,
we were driven by two different goals: high accuracy
and low inference execution time to maintain the soft
real-time property. To achieve the second goal we
heavily make use of the two following concepts:
• Convolutional layers with 1 × 1 filters: Strictly
speaking, those layers do not perform a con-
volution but combine the features of each pixel
densely to a new set of features while sharing the
trained weights over all pixels. In a CNN, some
classical convolutional layers can be replaced by
those layers to save execution time without losing
much accuracy.
• Feature reduction layers: As first applied in
the Inception Modules of the GoogleNet (Szegedy
et al., 2015), 1 × 1 convolution layers can be used
to reduce the number of features on each pixel be-
fore applying the next layer, which also has shown
Real-time Low SNR Signal Processing for Nanoparticle Analysis with Deep Neural Networks
39
Input Image Sequence - 8
2-Class Confidence Map
Conv. Layer 3x3 - 32
Max-Pooling - 32
Max-Pooling - 64
Conv. Layer 1x1 - 2
Upscaling x4
Fire Module – 8 / 64
Conv. 1x1 – Feature Reduce - 8
Conv. 3x3 - 32
Conv. 1x1 - 32
Filter Concatenation - 64
Fire Module – 8 / 64
Conv. 1x1 – Feature Reduce - 8
Conv. 3x3 - 32 Conv. 1x1 - 32
Filter Concatenation - 64
(a) Fully conv. detection network.
Conv. Layer 1x1 – 80
(Pixel-wise Fully-Connected)
Input Image Sequence - 32
Conv. Layer 1x1 - 64
(Pixel-wise Fully-Connected)
Conv. Layer 1x1 – 48
(Pixel-wise Fully-Connected)
Conv. Layer 1x1 – 32
(Pixel-wise Fully-Connected)
Conv. Layer 1x1 – 2
(Pixel-wise Fully-Connected)
2-Class Confidence Map
(b) Time-series analysis network.
Input Image Patch
2- Class Softmax Output
Conv. Layer 3x3 - 64
Max-Pooling - 64
Max-Pooling - 128
Conv. Layer 1x1 - 64
Fire Module – 16 / 128
Max-Pooling - 128
Fire Module – 16 / 128
Max-Pooling - 128
Fire Module – 16 / 128
Average-Pooling - 64
Fully-Connected - 2
(c) Particle classification network.
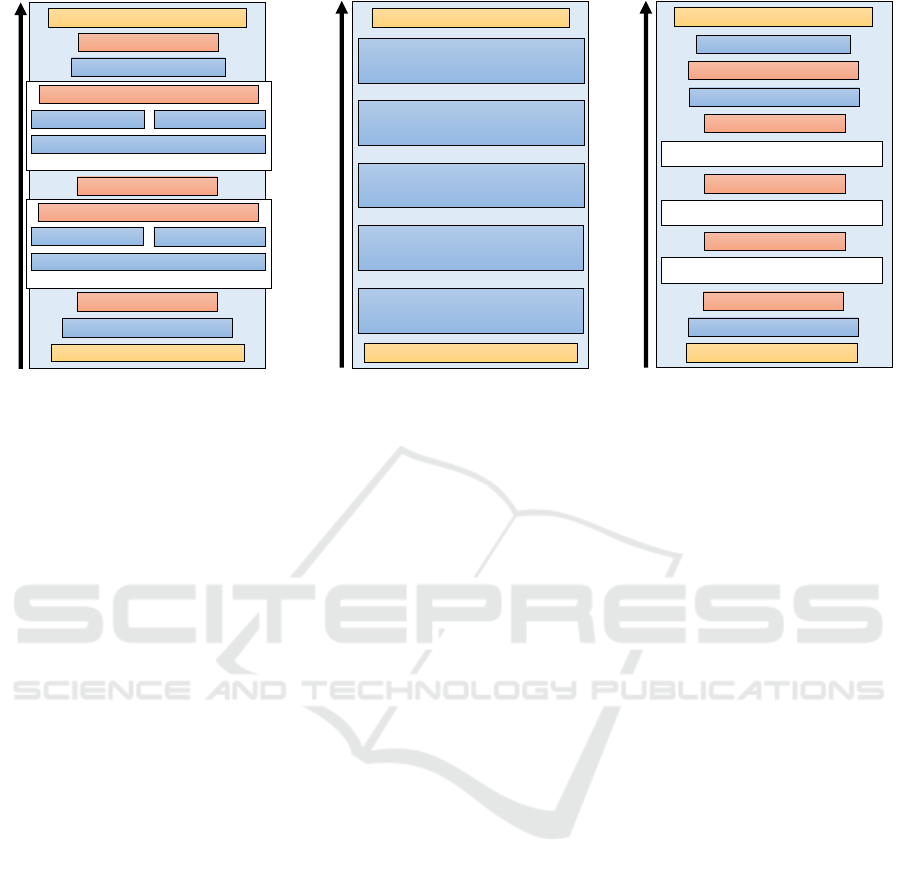
Figure 3: Architectures of our two different detection and our classification network. After the layer description, the number
of output feature maps is given for each layer. In each figure, the input is shown on the bottom and the output on the top.
to save execution time without sacrificing much
accuracy.
Fully-Convolutional Detection Network. The
first detection network combines the ideas of the
FCNs from Long et al. (Long et al., 2015) with the
efficiency of the inception modules of the GoogleNet
(Szegedy et al., 2015) and the spatial-temporal
early fusion method mentioned by Karpathy et al.
(Karpathy et al., 2014). The architecture is shown
in Figure 3a. As network input, we use a stack of 8
subsequent input images from the sensor data stream.
Then, the images are processed using scaled-down
inception modules, called fire modules, and max
pooling. The fire modules, as employed in the
SqueezeNet (Iandola et al., 2016), consist of three
convolutional layers. First the data is reduced by
applying a 1 ×1 convolutional layer. Then, the num-
ber of features is expanded again by another 1 ×1
and one 3 × 3 convolutional layer before both results
are concatenated along the feature dimension. After
three fire modules and max pooling layers, the down-
scaled feature maps are upscaled to input resolution
before computing the pixel-wise loss. As training
data, we use stacks of real sensor images together
with binary ground truth images that were automat-
ically derived from the manually created ground truth.
Time-Series Analysis Network. The second detec-
tion approach classifies the signal in a single pixel
over time. This has been accomplished with a 2-class,
5-layer MULTILAYER PERCEPTRON (MLP) classi-
fier. The input time-series consists of 32 signal values,
normalized to zero mean and a standard deviation of
one. Although this classification network is based on
an MLP architecture, it was realized using convolu-
tional layers, due to performance and practical rea-
sons on this particular application. The MLP classi-
fier realized as CNN is shown in Figure 3b. Since
the detection runs on an image sequence, the three-
dimensional inputs can be used as input for a CNN
with 1 ×1 convolutional layers, which yields a two-
dimensional feature map with the same width and
height as a single input image. Hence, every layer
in this CNN consists of n 1 × 1 filters, which is equiv-
alent to the densely connected layer with n outputs of
the MLP, applied on each pixel, individually.
The time-series classification network was trained
exclusively on synthesized data, which is motivated
by the fact that it is easier and more accurate to
generate realistic pixel time-series than it is to
manually label real data. The training data set
is composed of 5000 000 positive and 5000 000
negative training samples. The negative samples
consist of values drawn from a Gaussian distribution
N (µ, σ
2
), where means 0.1 ≤ µ ≤ 0.9 and standard
deviations 0.005 ≤ σ ≤ 0.25 are uniformly sampled
for each sample. For the positive examples, the same
procedure was used and an intensity step with step
height depending on target particle size was added on
top.
Particle Classification Network. We decided to
employ an independent classification CNN (LeCun
et al., 1995) after candidate generation to classify
between correct detections (particles) and false
detections (artifacts and noise). Since this network
BIOSIGNALS 2018 - 11th International Conference on Bio-inspired Systems and Signal Processing
40

is only applied on small signal patches, it allows the
application of a deeper network and more filters per
layer without destroying the real-time property. Our
classification architecture is displayed in Figure 3c.
It also makes use of SqueezeNet’s FireModules since
they have proven to be very fast and effective. As
input, the network receives 32 px × 32 px patches
while the output are confidences for two classes. The
network is trained using a set of signals that was
extracted from real sensor data, as described further
in Section 5 and shown in Figure 4.
Particle Size Estimation Network. The particle size
estimation is part of the previous work (Lenssen et al.,
2017) and is mentioned here for the sake of com-
pleteness. It consists of a CNN that simulates regres-
sion with classification through binning of the particle
sizes. It is trained using synthesized particle patches
containing averaged intensity peaks.
4.3 Frequency Domain Analysis
Frequency domain analysis has been used in the lit-
erature to detect the abnormalities in medical im-
ages (Aljarrah et al., 2015; Woodward et al., 2003).
We use it to compare our classification network to
a traditional approach on the same task. We extract
two types of frequency domain features, spectral and
wavelets features, to analyze the texture of the im-
age, because images that contain particles have dif-
ferent texture than images without particles, as shown
in Figure 4.
Figure 4: Example patches extracted from the sensor data.
The first three columns show signals of 200 nm, 100 nm and
80 nm particles, respectively. The fourth column displays
patches containing only noise and artifact signals without
particles. The images have been manually enhanced for vi-
sualization.
Spectral features can be used to characterize the
periodicity of the texture pattern by observing the
bursts in Fourier spectrum of the image. The features
include the peak value and its location, the mean, the
variance, and the distance between the mean and the
peak value of the spectrum (Gonzalez and Woods,
2006). Wavelet transform analysis is also used to de-
tect the particles by studying the frequency content of
the image in different scales. We use the Haar wavelet
transform with 3 scales to produce the coefficients for
10 channels. The energy values of the channels rep-
resent the texture features, which can be extracted by
calculating the mean magnitude of each channel’s co-
efficients as follows:
E =
1
M · N
M
∑
i=1
N
∑
j=1
| w(i, j) |, (3)
where M and N are the dimensions of the channel,
and w(i, j) is the wavelet coefficient (Castellano et al.,
2004). To classify the patches based on the extracted
features a Random Forest classifier was used.
4.4 Implementation Details
The following section presents details and parameters
of the training of neural networks, the Random Forest
classifiers and the pipeline implementations we used
to obtain our results.
Neural Network Training. The FCN and CNNs
were trained with TensorFlow (Abadi et al., 2015)
using the backpropagation algorithm (Hecht-Nielsen
et al., 1988) for gradient estimation and the Adam
optimization method (Kingma and Ba, 2014) with
an initial step size α = 0.001, exponential decay
rates β
1
= 0.9 and β
2
= 0.999, and ε = 10
−8
. In
contrast, the time-series analysis network was trained
with the ADAGRAD optimization method (Duchi
et al., 2011) with an initial step of α = 0.01 and an
exponential decay rate of 0.9. As mini-batch sizes,
we used 16 for the FCN detector and 256 for the
classification networks. The parameter settings were
chosen empirically based on preliminary work.
Training Data Augmentation. In addition to
dropout, we applied different data augmentation
techniques to further reduce overfitting. For the
input of the FCN detector and the CNN classifier
we applied small random intensity and contrast
modifications as well as random flipping. Intensity
and contrast modifications are always applied on the
whole image so that relative intensity information is
preserved. In addition to that, the 32 px × 32 px input
patches for classification are randomly cropped out
of 48 px × 48 px images. It should be noted that the
intensity input of the size estimation network is not
Real-time Low SNR Signal Processing for Nanoparticle Analysis with Deep Neural Networks
41

Table 1: Generated patches and captured image sequences used for evaluation with average particle size, number of
patches/frames, number of particles and median SNR.
Particle classification data sets Avg. part. size # Training images # Testing images
Ds1
Patches
100&200nm
100 nm & 200 nm 19344 19344
Ds2
Patches
80nm
80 nm 8308 8308
Ds3
Patches
80nm
80 nm 12916 3700
Sensor experiment data sets Avg. part. size # Frames # Part. Median SNR
Ds4
200nm
200 nm 2000 93 2.125
Ds5
100nm
100 nm 4000 56 1.247
Ds6
80nm
80 nm 6300 819 0.761
Ds7
80nm
80 nm 6750 214 0.716
Ds8
80nm
80 nm 4400 196 0.639
modified, since the absolute intensity holds important
information about particle size.
Random Forest Classifier. The Random Forest
classifier model has been trained with Weka (Hall
et al., 2009). A cross validation parameter tun-
ing (Kohavi, 1995) has been performed to optimize
the hyper-parameters of the Random Forest model.
The maximum depth of the trees has been optimized
from 5 to 20, the number of trees from 100 to 500,
and the number of random features from 4 to 9.
Image Processing Pipeline. The GPGPU image pro-
cessing pipeline, called VIRUS DETECTION WITH
OPENCL (VirusDetectionCL), was described and im-
plemented by Libuschewski using OPEN COMPUT-
ING LANGUAGE (OpenCL) (Libuschewski, 2017).
For real-time neural network inference, we use
DEEP RESOURCE-AWARE OPENCL INFERENCE
NETWORKS (deepRacin), an OpenCL-based neural
network inference library we created. Using this li-
brary, we are able to directly integrate our trained
networks into the VirusDetectionCL pipeline and ex-
ecute them on OpenCL-capable mobile and desktop
devices, utilizing the parallel processing power of
GPUs.
5 EXPERIMENTS
We provide results for two different types of experi-
ments. First, we solely evaluate the classification net-
work using dedicated classification benchmark data
sets, for which examples were given in Figure 4. Sub-
sequently, we use the whole pipeline, including de-
tection and classification networks, to analyze sensor
data.
The main focus is on the 80 nm data sets, which
have very low SNR. The SNR(S, B) for a given par-
ticle signal S and a given background signal B is cal-
culated according to the definition of Cheezum et al.
(Cheezum et al., 2001) as
SNR(S, B) =
|µ(S) − µ(B)|
σ(S)
, (4)
where S is a multiset of signal values, B a multiset
of background values, µ(S) the average of S, µ(B) the
average of B and σ(S) the standard deviation of S.
5.1 Data Set Acquisition
For capturing nanoparticles with the PAMONO sen-
sor we used glass slides covered with a layer of about
50 nm thickness encompassing 5 nm Titanium and ap-
proximately 45 nm gold (PHASIS, Switzerland) for
the sensor surface. A liquid containing around 10 %
of aluminum hydroxide (N
¨
uscoflock, Dr. N
¨
usgen
Chemie, Germany) was employed to cover the gold
layer. These gold bearing glass slides were attached
to the glass prism with a help of an immersion liq-
uid possessing the same refractive index as the prism.
A diode laser (HL6750MG, Thorlabs, Germany) pro-
vided an incidence beam with a wavelength λ ≈
675 nm for illumination of the gold layer through the
prism. A 50 mm photographic lens (Rokkor MD, Mi-
nolta, Japan) imaged the gold surface onto a 5 Mpx
camera (GC2450 Prosilica, Allied Vision, Germany).
Polystyrene nanoparticles (Molecular Probes, Life
Technologies, USA) of 200 nm, 100 nm and 80 nm
were pumped through the U-shaped flow cell as a
suspension in distilled water containing 0.3 % sodium
chloride. Image recording speed was 41 fps to 45 fps,
but was kept constant during each experiment. For
each suspension of 80 nm, 100 nm and 200 nm par-
ticles, image sequences were captured, picturing the
sensor surface.
BIOSIGNALS 2018 - 11th International Conference on Bio-inspired Systems and Signal Processing
42
The resulting data sets are listed in Table 1. The
first three data sets, named Ds1
Patches
100&200nm
, Ds2
Patches
80nm
and Ds3
Patches
80nm
contain extracted patches from the
recorded signal, cf. Figure 4, and the subsequent
five data sets, named Ds4
200nm
, Ds5
100nm
, Ds6
80nm
,
Ds7
80nm
and Ds8
80nm
are raw sensor image sequences
as recorded by the sensor. For all data sets, manually
created ground truths are available in which all parti-
cle signals were marked by human annotators.
5.2 Standalone Classification
Experiment
The first three data sets in Table 1 are benchmark
data sets for particle classification. They are used
to evaluate the classification CNN and the frequency
domain analysis and consist of 48 px × 48 px particle
signal patches, which were extracted from sensor sig-
nal. Data set Ds1
Patches
100&200nm
contains disjoint training
and testing patches extracted from data sets Ds4
200nm
and Ds5
100nm
. Data sets Ds2
Patches
80nm
and Ds3
Patches
80nm
contain patches from data sets Ds6
80nm
and Ds7
80nm
.
For data set Ds2
Patches
80nm
training and testing images are
drawn from both source data sets while for Ds3
Patches
80nm
,
training images are only extracted from Ds6
80nm
and
testing images from Ds7
80nm
, thus allowing to evalu-
ate the transferability of the model between different
measurements. In all 3 generated data sets, training
and testing data are disjoint and class-balanced, al-
lowing to use accuracy as quality measure.
We applied two different methods on these data
sets: the CNN classifier called M
CNN-Cl
and the fre-
quency domain analysis with Random Forest classi-
fier called M
FDA
RF
.
5.3 Sensor Data Experiment
We evaluate the whole processing pipeline by com-
paring the results (proposed particles that were posi-
tively classified) to the manually created ground truth.
Data set Ds6
80nm
was used for training the detector
and classification network. The trained models were
tested on data sets Ds4
200nm
, Ds5
100nm
, Ds7
80nm
and
Ds8
80nm
.
As quality measures, precision, recall and the F
1
-
score (Powers, 2011) are used. The F
1
-score is de-
fined as a balance between precision and recall:
F
1
=
2 · precision · recall
precision + recall
. (5)
We differentiate between results obtained by apply-
ing the detection step only and that obtained by de-
tecting and classifying. Ideally, the detector provides
high recall while the classification is able to sort out
false positives without sorting out too many true pos-
itives. Therefore, we applied and compared five dif-
ferent methods:
• M
Baseline
: baseline pipeline (previous work),
• M1
FCN-Det
: the FCN detector without classifica-
tion,
• M2
FCN-Det
CNN-Cl
: the FCN detector with CNN classifi-
cation,
• M3
TS-Det
: the time-series analysis network with-
out classification,
• M4
TS-Det
CNN-Cl
: the time-series detection network with
CNN classification.
Ds1
P atches
100&200nm
Ds2
P atches
80nm
Ds3
P atches
80nm
0
0.2
0.4
0.6
0.8
1
Precision / Recall
M
F DA
RF
: Precision
M
CN N −C l
: Precision
M
F DA
RF
: Recall
M
CN N −C l
: Recall
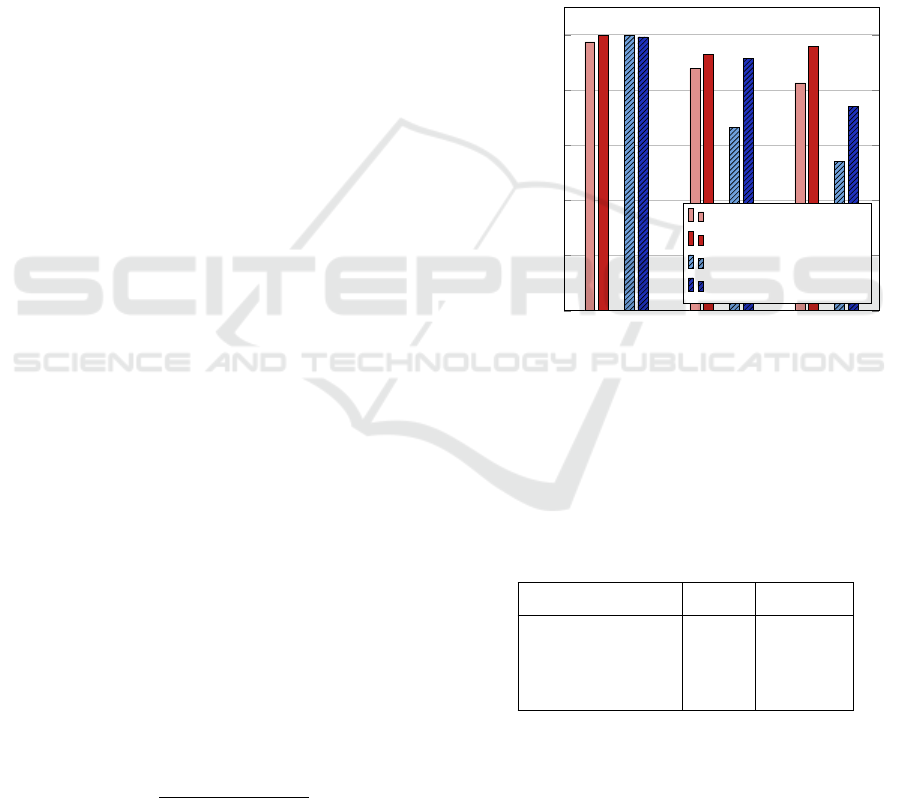
Figure 5: Precision and recall of the classification results.
Values are given for the positive class. Mostly, precision
is higher than recall, as preferred. The M
CNN-Cl
method
provides stronger results.
Table 2: Classification accuracy results for the generated
data sets and the two methods: frequency domain analysis
plus Random Forest classifier M
FDA&RF
and CNN classifi-
cation M
CNN-Cl
.For each row, the bold value shows the best
result.
Data set / method M
FDA
RF
M
CNN-Cl
Ds1
Patches
100&200nm
0.985 0.995
Ds2
Patches
80nm
0.786 0.922
Ds3
Patches
80nm
0.713 0.854
6 RESULTS AND DISCUSSION
In the following Sections 6.1 and 6.2, we present and
discuss results for the two different classes of experi-
ments that were described in Section 5. Then, we pro-
vide benchmark results for our pipeline in Section 6.3.
Real-time Low SNR Signal Processing for Nanoparticle Analysis with Deep Neural Networks
43
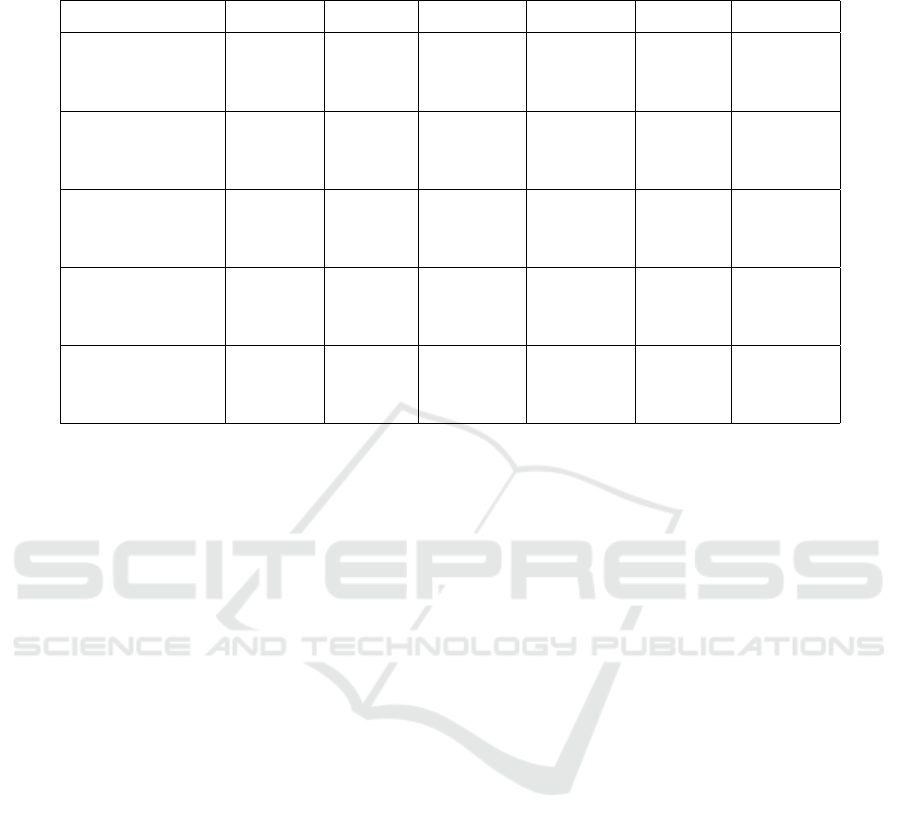
Table 3: Detection results evaluated for the baseline method and the four presented methods on five data sets. Quality measures
are given as precision, recall and F
1
-score. For each row, the bold value shows the best result.
Data set / method M
Baseline
M1
FCN-Det
M2
FCN-Det
CNN-Cl
M3
TS-Det
M4
TS-Det
CNN-Cl
Ds4
200nm
Precision 0.909 0.888 0.908 0.933 0.933
Recall 0.787 0.763 0.742 0.903 0.892
F
1
-score 0.844 0.821 0.817 0.918 0.918
Ds5
100nm
Precision 0.769 0.814 0.842 0.235 0.938
Recall 0.798 0.857 0.857 0.929 0.803
F
1
-score 0.783 0.835 0.810 0.375 0.865
Ds6
80nm
Precision 0.410 0.782 0.877 0.098 0.887
Recall 0.492 0.715 0.677 0.967 0.506
F
1
-score 0.448 0.747 0.764 0.177 0.644
Ds7
80nm
Precision 0.330 0.347 0.840 0.025 0.829
Recall 0.549 0.795 0.707 0.967 0.428
F
1
-score 0.412 0.483 0.768 0.049 0.564
Ds8
80nm
Precision 0.053 0.258 0.801 0.017 0.729
Recall 0.561 0.782 0.553 0.970 0.355
F
1
-score 0.097 0.388 0.655 0.033 0.478
6.1 Standalone Particle Classification
The results for the standalone particle classification
are shown in Table 2. It shows classification accuracy
for the two presented classification methods M
FDA
RF
and M
CNN-Cl
.
We observe nearly the same accuracy of both
motehods on the easier Ds1
Patches
100&200nm
data set. How-
ever, the results on the 80 nm data sets Ds2
Patches
80nm
and
Ds3
Patches
80nm
show that the M
CNN-Cl
method outperforms
the M
FDA
RF
method on harder tasks. The difference in
accuracy between data sets Ds2
Patches
80nm
and Ds3
Patches
80nm
show that transferring the model to a different mea-
surement is possible but leads, in this case, to a sig-
nificant decrease in accuracy. However, we show that
even the transferred classification model is able to im-
prove the detection results provided in the next sec-
tion.
For more insight, we also detail precision and re-
call of each approach and data set in Figure 5. In
general, the precision is higher than the recall, which
is also what is preferred most of the time. In addi-
tion, the M
CNN-Cl
method shows better results than
the M
FDA
RF
method in both criteria. All in all, these re-
sults led us to the decision to use the M
CNN-Cl
method
together with our detection networks for our sensor
data experiments that are presented in the following
section.
6.2 Sensor Data Experiment
The results for the sensor data experiment are shown
in Table 3. The quality measures precision, recall and
F
1
-score are provided for the baseline method and the
four presented methods on five data sets. The best
results of each experiment are printed bold.
First, it should be noted that Ds6
80nm
was used to
train the M1
FCN-Det
detector and to extract the training
data for data set Ds3
Patches
80nm
, which was used to train
the applied M
CNN-Cl
classifier. Therefore, the results
of methods M1
FCN-Det
, M2
FCN-Det
CNN-Cl
and M4
TS-Det
CNN-Cl
for
Ds6
80nm
are training scores. The other data sets were
not seen during training and used to calculate the test-
ing score. Since the time-series classification network
was trained using synthetic data, all results of method
M3
TS-Det
are test results.
The evaluation shows that nearly all our methods
outperform the baseline method M
Baseline
by a large
margin. The M3
TS-Det
and M4
TS-Det
CNN-Cl
methods provide
strong results on data sets with a median SNR above
one. For data set Ds4
200nm
, the classifier is not even
needed to sort out false positives. For data sets with a
median SNR below one however, the resulting time-
series detection network shows very high sensitivity,
resulting in low precision in order to find most true
positives. Therefore, it heavily relies on the classifier
to sort out false positives consisting of noise and arti-
facts. For these data sets, the combination of the FCN
detector and the CNN classifier, method M2
FCN-Det
CNN-Cl
,
proves to be the strongest method. It achieves a preci-
sion above 0.8 on all three data sets, thus having a low
BIOSIGNALS 2018 - 11th International Conference on Bio-inspired Systems and Signal Processing
44

number of false positives. This indicates that, espe-
cially for low SNR signals, the local spatial informa-
tion is important to perform reliable detection and that
time-series information of one pixel is not enough to
distinguish between artifact and particle signals. The
recall, despite being not optimal, is sufficient for a
lot of tasks, in which the existence and size distribu-
tions of particles should be derived. The worse results
for this method on data sets Ds4
200nm
and Ds5
100nm
is
easily explained by the fact that the networks were not
trained with images containing 100 nm and 200 nm
particles.
6.3 Performance Analysis
We profiled the proposed processing pipeline as well
as each deep neural network on an NVIDIA GeForce
GTX 1080 Ti. For whole pipeline application, we
achieve 15.296 ms per frame (65.4 fps) when using
the M4
TS-Det
CNN-Cl
method and 23.478 ms (42.5 fps) when
using the M4
TS-Det
CNN-Cl
method. Applying the FCN de-
tector on one image takes 0.827 ms while the time-
series analysis network is slower and takes 9.675 ms.
The classification CNN takes 0.119 ms per patch clas-
sification. All measurements were performed on data
set Ds7
80nm
, which has 6750 images with a resolution
of 880 px × 115 px.
7 CONCLUSIONS
In this work, we proposed additional deep neural net-
work methods for the PAMONO sensor data analy-
sis pipeline. We showed that through this extensions,
the detection of blobs with a median SNR below one
is possible. All in all, we achieved results on sig-
nals with a median SNR of 0.7 that were previously
reached on data sets with a median SNR of 1.25.
Our pipeline, consisting of detection and classifi-
cation networks, was able to achieve sufficiently high
results for most real world nanoparticle analysis ap-
plications of the PAMONO sensor while fulfilling the
soft real-time property on desktop GPUs. We are able
to successfully analyze suspensions containing 80 nm
polystyrene particles, given our current sensor experi-
ment setup. Detecting even smaller particles in the fu-
ture requires either the improvement of the data (with
higher SNR for the same particle size) or the capa-
bility of the detection pipeline to handle even lower
SNR. In addition, we aim to bring the pipeline to
embedded GPUs, to further move towards the goal
of small, mobile nanoparticle analysis with the PA-
MONO sensor.
ACKNOWLEDGMENTS
This work has been supported by DEUTSCHE
FORSCHUNGSGEMEINSCHAFT (DFG) within the
Collaborative Research Center SFB 876 Providing In-
formation by Resource-Constrained Analysis, project
B2.
REFERENCES
Abadi, M., Agarwal, A., Barham, P., Brevdo, E., Chen, Z.,
Citro, C., Corrado, G. S., Davis, A., Dean, J., Devin,
M., Ghemawat, S., Goodfellow, I., Harp, A., Irving,
G., Isard, M., Jia, Y., Jozefowicz, R., Kaiser, L., Kud-
lur, M., Levenberg, J., Man
´
e, D., Monga, R., Moore,
S., Murray, D., Olah, C., Schuster, M., Shlens, J.,
Steiner, B., Sutskever, I., Talwar, K., Tucker, P., Van-
houcke, V., Vasudevan, V., Vi
´
egas, F., Vinyals, O.,
Warden, P., Wattenberg, M., Wicke, M., Yu, Y., and
Zheng, X. (2015). Tensorflow: Large-scale machine
learning on heterogeneous systems.
Aljarrah, I., Toma, A., and Al-Rousan, M. (2015). An au-
tomatic intelligent system for diagnosis and confirma-
tion of johne’s disease. Int. J. Intell. Syst. Technol.
Appl., 14(2):128–144.
Beusink, J. B., Lokate, A. M. C., Besselink, G. A. J., Pruijn,
G. J. M., and Schasfoort, R. B. M. (2008). Angle-
scanning spr imaging for detection of biomolecular
interactions on microarrays. Biosensors and Bioelec-
tronics, 23(6):839–844.
Breiman, L. (2001). Random forests. Machine Learning,
45(1):5–32.
Castellano, G., Bonilha, L., Li, L. M., and Cendes, F.
(2004). Texture analysis of medical images. Clinical
Radiology, 59(12):1061–1069.
Cheezum, M. K., Walker, W. F., and Guilford, W. H. (2001).
Quantitative comparison of algorithms for tracking
single fluorescent particles. Biophysical Journal,
81(4):2378–2388.
Chinowsky, T. M., Mactutis, T., Fu, E., and Yager, P. (2004).
Optical and electronic design for a high-performance
surface plasmon resonance imager. In Optical Tech-
nologies for Industrial, Environmental, and Biological
Sensing, pages 173–182.
Dragovic, R. A., Gardiner, C., Brooks, A. S., Tannetta,
D. S., Ferguson, D. J., Hole, P., Carr, B., Redman,
C. W., Harris, A. L., Dobson, P. J., Harrison, P.,
and Sargent, I. L. (2011). Sizing and phenotyping
of cellular vesicles using nanoparticle tracking anal-
ysis. Nanomedicine: Nanotechnology, Biology and
Medicine, 7(6):780 – 788.
Duchi, J., Hazan, E., and Singer, Y. (2011). Adaptive sub-
gradient methods for online learning and stochastic
optimization. Journal of Machine Learning Research,
12(Jul):2121–2159.
Dulbecco, R. (1954). Plaque formation and isolation of pure
lines with poliomylitis viruses. Journal of Experimen-
tal Medicine, 99(2):167–182.
Real-time Low SNR Signal Processing for Nanoparticle Analysis with Deep Neural Networks
45

Gan, S. D. and Patel, K. R. (2013). Enzyme immunoassay
and enzyme-linked immunosorbent assay. The Jour-
nal of Investigative Dermatology, 133(9):1–3.
Giebel, K.-F., Bechinger, C. S., Herminghaus, S., Riedel,
M., Leiderer, P., Weiland, U. M., and Bastmeyer, M.
(1999). Imaging of Cell/Substrate Contacts of Living
Cells with Surface Plasmon Resonance Microscopy.
Bibliothek der Universit
¨
at Konstanz, Konstanz, Ger-
many.
Gonzalez, R. C. and Woods, R. E. (2006). Digital Image
Processing (3rd Edition). Prentice-Hall, Inc, Upper
Saddle River, NJ, USA.
Hall, M., Frank, E., Holmes, G., Pfahringer, B., Reutemann,
P., and Witten, I. H. (2009). The weka data mining
software: An update. ACM SIGKDD Explorations
Newsletter, 11(1):10–18.
Hecht-Nielsen, R. et al. (1988). Theory of the backpropaga-
tion neural network. Neural Networks, 1(Supplement-
1):445–448.
Iandola, F. N., Moskewicz, M. W., Ashraf, K., Han, S.,
Dally, W. J., and Keutzer, K. (2016). Squeezenet:
Alexnet-level accuracy with 50x fewer parameters and
<1mb model size. CoRR, abs/1602.07360.
Karpathy, A., Toderici, G., Shetty, S., Leung, T., Suk-
thankar, R., and Li, F.-F. (2014). Large-scale video
classification with convolutional neural networks. In
2014 IEEE Conference on Computer Vision and Pat-
tern Recognition, CVPR 2014, Columbus, OH, USA,
June 23-28, 2014, pages 1725–1732.
Kingma, D. P. and Ba, J. (2014). Adam: A method for
stochastic optimization. In Proceedings of the 3rd In-
ternational Conference on Learning Representations
(ICLR).
Kohavi, R. (1995). Wrappers for Performance Enhance-
ment and Oblivious Decision Graphs. PhD thesis,
Stanford University, Department of Computer Sci-
ence, Stanford University.
Kretschmann, E. (1971). The determination of the optical
constants of metals by excitation of surface plasmons.
European Physical Journal A, 241(4):313–324.
LeCun, Y., Bengio, Y., et al. (1995). Convolutional net-
works for images, speech, and time series. The
handbook of brain theory and neural networks,
3361(10):1995.
Lenssen, J. E., Shpacovitch, V., and Weichert, F. (2017).
Real-time virus size classification using surface plas-
mon pamono resonance and convolutional neural net-
works. In Maier-Hein, K. H., Deserno, T. M., Han-
dels, H., and Tolxdorff, T., editors, Bildverarbeitung
f
¨
ur die Medizin 2017, Informatik Aktuell, pages 98–
103. Springer, Berlin, Germany and Heidelberg, Ger-
many.
Libuschewski, P. (2017). Exploration of Cyber-Physical
Systems for GPGPU Computer Vision-Based Detec-
tion of Biological Viruses . PhD thesis, TU Dortmund,
Dortmund, Germany.
Liu, J., White, J. M., and Summers, R. M. (2010). Auto-
mated detection of blob structures by hessian analysis
and object scale. In Image Processing (ICIP). 17th
IEEE International Conference on, pages 841–844.
Long, J., Shelhamer, E., and Darrell, T. (2015). Fully con-
volutional networks for semantic segmentation. In
IEEE Conference on Computer Vision and Pattern
Recognition, CVPR 2015, Boston, MA, USA, June 7-
12, 2015, pages 3431–3440.
Moon, W. K., Shen, Y.-W., Bae, M. S., Huang, C.-S., Chen,
J.-H., and Chang, R.-F. (2013). Computer-aided tumor
detection based on multi-scale blob detection algo-
rithm in automated breast ultrasound images. Medical
Imaging. IEEE Transactions on, 32(7):1191–1200.
Naimushin, A. N., Soelberg, S. D., Bartholomew, D. U.,
Elkind, J. L., and Furlong, C. E. (2003). A portable
surface plasmon resonance (spr) sensor system with
temperature regulation. Sensors and Actuators B:
Chemical, 96(1-2):253–260.
Pattnaik, P. (2005). Surface plasmon resonance: Applica-
tions in understanding receptor-ligand interaction. Ap-
plied Biochemistry and Biotechnology, 126(2):79–92.
Powers, D. M. W. (2011). Evaluation: From precision, re-
call and f-measure to roc., informedness, markedness
& correlation. Journal of Machine Learning Tech-
nologies, 2(1):37–63.
Scarano, S., Ermini, M. L., Mascini, M., and Minunni,
M. (2011). Surface plasmon resonance imaging for
affinity-based sensing: An analytical approach. In
BioPhotonics. International Workshop on, pages 957–
966.
Shpacovitch, V., Temchura, V., Matrosovich, M.,
Hamacher, J., Skolnik, J., Libuschewski, P., Siedhoff,
D., Weichert, F., Marwedel, P., M
¨
uller, H.,
¨
Uberla, K.,
Hergenr
¨
oder, R., and Zybin, A. (2015). Application
of surface plasmon resonance imaging technique for
the detection of single spherical biological submi-
crometer particles. Analytical Biochemistry: Methods
in the Biological Sciences, 486:62–69.
Siedhoff, D. (2016). A parameter-optimizing model-based
approach to the analysis of low-snr image sequences
for biological virus detection. phd, Universit
¨
at Dort-
mund. Publikation.
Siedhoff, D., Libuschewski, P., Weichert, F., Zybin, A.,
Marwedel, P., and M
¨
uller, H. (2014). Modellierung
und optimierung eines biosensors zur detektion viraler
strukturen. In Deserno, T. M., Handels, H., Meinzer,
H.-P., and Tolxdorff, T., editors, Bildverarbeitung f
¨
ur
die Medizin (BVM), Informatik Aktuell, pages 108–
113. Springer, Berlin, Germany and Heidelberg, Ger-
many.
Smal, I., Loog, M., Niessen, W., and Meijering, E. (2009).
Quantitative comparison of spot detection methods
in live-cell fluorescence microscopy imaging. In
Biomedical Imaging: From Nano to Macro. IEEE In-
ternational Symposium on, pages 1178–1181.
Steiner, G. and Salzer, R. (2001). Biosensors based on spr
imaging. In T
¨
ubingen, U., editor, 2. BioSensor Sym-
posium, T
¨
ubingen, Germany. Universit
¨
at T
¨
ubingen.
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S.,
Anguelov, D., Erhan, D., Vanhoucke, V., and Rabi-
novich, A. (2015). Going deeper with convolutions.
In IEEE Conference on Computer Vision and Pattern
Recognition (CVPR), pages 1–9. IEEE.
BIOSIGNALS 2018 - 11th International Conference on Bio-inspired Systems and Signal Processing
46

Woodward, R. M., Wallace, V. P., Arnone, D. D., Linfield,
E. H., and Pepper, M. (2003). Terahertz pulsed imag-
ing of skin cancer in the time and frequency domain.
Journal of Biological Physics, 29(2-3):257–259.
Zybin, A. (2010). DE patent 10,2009,003,548 A1: Ver-
fahren zur hochaufgel
¨
osten erfassung von nanopar-
tikeln auf zweidimensionalen messfl
¨
achen.
Zybin, A. (2013). US patent 8,587,786 B2: Method
for high-resolution detection of nanoparticles on two-
dimensional detector surfaces.
Real-time Low SNR Signal Processing for Nanoparticle Analysis with Deep Neural Networks
47
