
A Methodology to Perform Semi-automatic Distributed EHR Database
Queries
Olga Fajarda
1
, Lu
´
ıs Basti
˜
ao Silva
2
, Peter R. Rijnbeek
3
,
Michel Van Speybroeck
4
and Jos
´
e Lu
´
ıs Oliveira
1
1
University of Aveiro, DETI/IEETA, Portugal
2
BMD Software, Aveiro, Portugal
3
Erasmus MC, Rotterdam, The Netherlands
4
Janssen Pharmaceutica NV, Beerse, Belgium
Keywords:
Electronic Health Records, Observational Studies, Cohorts, Clinical Research, Secondary Use.
Abstract:
The proliferation of electronic health databases has resulted in the existence of a wide collection of diversified
clinical digital data. These data are fragmented over dispersed databases in different clinical silos around the
world. The exploration of these electronic health records (EHRs) is essential for clinical and pharmaceutical
research and, therefore, solutions for secure sharing of information across different databases are needed.
Although several partial solutions have been proposed over the years, data sharing and integration has been
hindered by many ethical, legal and social issues. In this paper, we present a methodology to perform semi-
automatic queries over longitudinal clinical data repositories, where every data custodian maintains full control
of data.
1 INTRODUCTION
Over the past two decades the use of electronic health
record (EHR) systems has significantly increased in
many countries around the world. This increase
has resulted in the proliferation of electronic health
databases containing a wide collection of diversified
clinical digital data. Beyond the undeniable value
that EHRs have for the direct health care of patients,
i.e. beyond primary clinical care, the secondary use
of these data brings great benefit to scientific, clin-
ical and translational research. It can improve the
quality of healthcare services, allow public health and
pharmaceutical surveillance, monitor health crises,
increase the understanding of diseases, and can lead
to the development of new treatments (Hersh, 2007;
Safran et al., 2007).
The secondary use of clinical data opens the door
to translational research, which can be considered
a two-way path. The first is from “Bench to Bed-
side”, that is, translate research discoveries into clin-
ical practice; the second is from “Bedside to Bench”,
i.e., the other direction, using clinical practice to as-
sist research. The reuse of clinical digital data is very
useful in both ways, allowing time-saving and cost re-
duction, and avoiding redundant data collection.
In the development of new therapies, the sec-
ondary use of clinical digital data can improve the
clinical trial design and accelerate the complex pro-
cess of identifying clinical trial participants (Ohmann
and Kuchinke, 2007; Pakhomov et al., 2007). A feasi-
bility trial usually starts by asking data custodians or
physicians if they have patients who meet research el-
igibility criteria. For a clinical trial to be scientifically
and statistically valid, the number of participants must
be sufficiently large (K
¨
opcke and Prokosch, 2014),
and so this process can be very slow and expensive.
The use of EHR data can reduce the time and cost of
this process. Besides, a pre-trial feasibility analysis
using EHR data also allows a redefinition of criteria
in order to increase the number of participants (Doods
et al., 2014).
Some adverse drug events are only observed after
its release to a large and diverse population because
a clinical trial has only a reduced number of partic-
ipants. Drug safety surveillance, that is, monitoring
medical product safety, can be done using EHRs (Tri-
fir
`
o et al., 2014).
EHRs can also be reused to conduct observational
studies, such as retrospective cohort studies and case-
control studies. A cohort study is a form of longi-
tudinal study used to study the incidence, causes, and
Fajarda, O., Silva, L., Rijnbeek, P., Speybroeck, M. and Oliveira, J.
A Methodology to Perform Semi-automatic Distributed EHR Database Queries.
DOI: 10.5220/0006579701270134
In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2018) - Volume 5: HEALTHINF, pages 127-134
ISBN: 978-989-758-281-3
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
127

prognosis of a given clinical condition. In a retrospec-
tive cohort study, one or more groups of patients are
followed up backwards to examine medical events or
outcomes (Mann, 2003). Some authors used EHRs
to do retrospective cohort studies, but they only used
EHRs collected in a few healthcare centers (Harris
et al., 2010; McDonald et al., 2014; Reisner et al.,
2015). In case-control studies, two groups of people,
one with the outcome of interest and the other with-
out it, are compared, retrospectively, on the basis of
the exposure to some agent or treatment (Song and
Chung, 2010) and, once again, this comparison can
be done using EHRs.
Despite the recognized value of the secondary use
of EHR, it is still nearly impossible to obtain access to
digital clinical data. Lopes et al. (Lopes et al., 2015)
reviewed initiatives and projects focusing on the ex-
ploration of patient-level data and pointed out that
even data obtained through public research funding
projects are not shared with the research community.
There are several reasons for this difficulty of
sharing patient-level digital data. One impediment
is the existence of database silos. Over the years, as
clinical digital data were collected in different coun-
tries and institutions, many isolated silos were created
due to the lack of regulation and primitive technolog-
ical implementation (Lopes et al., 2015; Miller and
Tucker, 2014). Due to these database silos, it is dif-
ficult for many researchers to locate the appropriate
dataset needed for their studies.
Clinical digital data are also widely distributed
and fragmented. A patient’s clinical history may be
fragmented and distributed among multiple electronic
systems, such as the patient’s pharmacy, insurance
companies, care providers and others (Pringle and
Lippitt, 2009). These distributed, decentralized and
autonomous EHR systems lead to the existence of
multiple highly heterogeneous databases, since every
system collects and stores the data in an application-
specific or vendor-specific format without consider-
ing information sharing. The heterogeneity of the
databases can be found at several levels, namely, in
the technologies and data models employed, in the
query languages supported and the terminologies rec-
ognized.
Another major impediment relates to privacy is-
sues due to legal, ethical and regulatory require-
ments (Cushman et al., 2010). Data privacy protec-
tion is a very important and sensitive matter because
a minimal break in privacy can have dramatic con-
sequences for individuals’ lives, healthcare providers
and subgroups within society. Moreover, legislation
differs from one country to another and it may be dif-
ficult to develop a protocol that conforms to all of
them (Meystre et al., 2017). The upcoming EU Gen-
eral Data Protection Regulation (GDPR)
1
will hope-
fully address this caveat.
For the success of clinical translational research,
it is imperative to develop solutions that enable
the querying of distributed and heterogeneous EHR
databases without losing data and patients’ privacy.
This paper presents a methodology to semi-
automatically query several distributed, heteroge-
neous databases. In Section 2 we present an overview
of existing solutions, while our methodology is pre-
sented in section 3. In Section 4 we discuss a proof-
of-concept implementation and Section 5 concludes
the paper.
2 RELATED WORK
As awareness of the value of secondary use of EHR
increased, several projects emerged to develop solu-
tions for secure sharing of information across dif-
ferent databases. These solutions have been funda-
mentally guided by two distinct approaches: they
can be centralized, where EHRs of dispersed systems
are copied and integrated into a single centralized
database, or they can be distributed, where the aim
is to query multiple physically distributed healthcare
data sources.
Mini-Sentinel (Platt and Carnahan, 2012) is a
project developed by the U.S. Food and Drug Admin-
istration (FDA) to perform active safety surveillance
of FDA-regulated medical products using routinely
collected electronic health record data from multiple
sources. The developed system uses data from pub-
lic and private organizations, centralized in a secure
container. A common data model was designed so
that each data partner is able to transform local source
data into this model. Several complementary software
tools have been developed to support specific research
questions, related to identification and evaluation of
the exposure of medical products and possible associ-
ated health issues. However, the setup of these tech-
nologies requires some technical expertise and field
knowledge.
The Informatics for Integrating Biology and the
Bedside (i2b2) (Murphy et al., 2010) is a U.S. project
launched with the aim to develop tools that can
help clinical researchers integrate medical records
and clinical research data in the genomics age. The
i2b2 team developed a web application which allows
cohort estimation and feasibility determination by
querying de-identified and aggregate EHR data. The
1
http://www.eugdpr.org/
HEALTHINF 2018 - 11th International Conference on Health Informatics
128

i2b2 team also developed the Shared Health Research
Information Network (SHRINE) (McMurry et al.,
2013; Weber et al., 2009), a distributed query sys-
tem that allows researchers to query synchronously
several databases containing everyday clinical data.
SHRINE provides obfuscated, aggregated counts of
patients, which facilitates population-based research
and assessment of potential clinical trial cohorts. The
software developed by the i2b2 is open source, freely
available and can be adapted to query other groups of
databases.
The Electronic Health Records for Clinical Re-
search (EHR4CR), was a European public-private
project that developed a platform to assist researchers
in clinical trials’ feasibility assessment and patient
recruitment (Daniel et al., 2016). Through a dis-
tributed real-time querying system, multiple clini-
cal data warehouses across Europe containing de-
identified EHR data, can be synchronously queried to
obtain aggregated results. The platform may enable
a trial sponsor to predict the number of eligible pa-
tients for a candidate clinical trial protocol, to assess
its feasibility and to locate the most relevant hospital
sites.
Another U.S. initiative is OHDSI (Observational
Health Data Sciences and Informatics) (Hripcsak
et al., 2015) a multi-stakeholder and interdisciplinary
project whose main purpose is to facilitate the anal-
yses of large-scale observational health data. This
worldwide initiative grew out of the Observational
Medical Outcomes Partnership (OMOP) and devel-
ops new solutions for data gathering and aggrega-
tion, promoting a standardized data model for patient-
level database representation, the OMOP Common
Data Model (OMOP CDM). Besides the common
data model, the OHDSI community has been develop-
ing several analytic tools, such as Achilles, Achilles
Web, HERMES and CIRCE. More recently, they de-
veloped a web-based platform, ATLAS, which in-
tegrates features from various previously developed
OHDSI applications. This platform allows database
exploration, standardized vocabulary browsing, co-
hort definition, and population-level analysis of obser-
vational data converted to the OMOP Common Data
Model.
The European Medical Information Frame-
work (EMIF)
2
is one of the most recent European
projects, aiming to facilitate the reuse and exploita-
tion of patient-level data from different EHR sys-
tems and cohorts, for research purposes (Lopes et al.,
2015). The EMIF Platform intends to be an in-
tegrated system to allow researchers to browse in-
formation at three different conceptual levels. The
2
http://www.emif.eu
first level refers to browsing a catalog containing
database fingerprints, i.e. a general characteriza-
tion of the databases (Basti
˜
ao et al., 2014), the sec-
ond level will allow the extraction of aggregated data
from several databases and the third level will allow
drilling down to the individual patient level in those
databases. EMIF has also adopted OMOP-CDM for
EHR data harmonization. Currently, the EMIF Cat-
alogue includes information from 6 research com-
munities, from population-based data sources (e.g.
electronic health records, regional databases) up to
disease-specific ones (e.g. Alzheimer).
Most of the solutions presented combine data
from healthcare centers which adopt the same data
model and allow the integration or distributed query
of databases. However, data sharing cannot be taken
for granted, and it might even be impossible for many
centers. Data custodians’ desire to share clinical data
for research is usually hindered by legal and gover-
nance issues, and they do not engage in solutions that,
for instance, use centralized data warehouses or real-
time query systems. Therefore, clinical research is
still hindered by the limited and fragmented access to
health data repositories. The methodology we present
allows clinical researchers to query several heteroge-
neous databases while keeping patient health data pri-
vate in each healthcare institution.
3 METHODOLOGY
The methodology we present enables semi-
automatically querying of several distributed,
heterogeneous EHR databases at once, which stream-
lines the entire request process. This approach is
semi-automatic so that every data custodian can
maintain control of their database and only share
the data they consider to fulfill the legal, ethical and
regulatory requirements. Moreover, the methodology
uses partially existing solutions and open-source soft-
ware, which significantly reduces the cost involved in
the process.
Our methodology has three main actors:
• the Researcher, the person who wants to query
one or several patient-level databases;
• the Data Custodian (DC), the person responsible
for managing a database;
• the Study Manager (SM), the person who leads
and manages the research study and moderates the
tasks between the researcher and the Data Custo-
dian.
Other actors can be involved in the process, e.g. the
A Methodology to Perform Semi-automatic Distributed EHR Database Queries
129

SM can delegate some of their tasks and responsibili-
ties to others.
The methodology assumes the use of a publicly
available common data model and an open-source an-
alytic tool that releases statistical and aggregated in-
formation on clinical digital data converted to this
model. Several authors (Kahn et al., 2012; Ogun-
yemi et al., 2013; Ross et al., 2014) compared some
of the existing common data models, including the
ones from OMOP, from Mini-Sentinel, and from i2b2,
and they all concluded that the OMOP CDM was the
most complete and efficient. Many data custodians
worldwide have already converted the data from their
databases to OMOP CDM. For instance, OHDSI Eu-
rope
3
is a recent initiative that aims to build a strong
European OHDSI community to actively contribute
to the implementation and further development of
OMOP-CDM and its analytical tools.
ATLAS
4
is the open-source web application used
to conduct scientific analyses on standardized obser-
vational data converted to the OMOP CDM. This an-
alytical tool allows the generation and execution of
scripts with cohort definitions, which considerably
simplifies the data custodians’ work when asked to
query their databases. Although another common
data model can be used, we assume in the rest of the
paper that all the databases involved in the process
were converted to the OMOP CDM and the analytical
tool used is ATLAS.
Our approach also assumes that the EMIF Cat-
alogue is the main entry and management solution,
where researchers can search for data sources, submit
a study request, choose the databases to engage with,
and follow the progress of the study, while others (SM
and DC) are handling the data extraction job. So all
communication between all users is through this ap-
plication. In addition, a workflow management tool is
used to perform and monitor all the tasks involved in
the process.
The SM manages the entire query process. They
receive all the study requests, evaluate their suitabil-
ity and also the DCs’ willingness to participate, cre-
ate an ATLAS script that defines the cohort, share it
with the DCs, and after receiving the DCs’ response,
reply to the study request. The SM is a community
expert that knows the characteristics of the different
databases that are part of the group, and is familiar
with the technologies and software needed to query
these databases, namely the EMIF Platform and AT-
LAS.
The DC is responsible for the local running of the
script sent by the SM and determines if the results of a
3
http://www.ohdsi-europe.org
4
http://www.ohdsi.org/web/atlas
query can be shared. Since this methodology does not
require all the data to be centralized, nor does it need
to previously de-identify the data, the DC keeps au-
tonomy and control of its database and the executing
and sharing of query results.
Figure 1 presents the main workflow of this
methodology. The researcher starts by formulating a
study request, which can be done by simply specify-
ing a question. This request is made using the EMIF
platform where the researcher also has access to a cat-
alogue of databases that can be chosen.
Afterwards, the SM analyses the study request and
decides if they can fulfill the request or if they need
more detailed information about the request in oder
to accurately define the cohort, in which case they
contact the researcher using the platform. The SM
can also make suggestions on how to formulate the
study request in order to be accepted. After accepting
the request, the SM uses a workflow management tool
to create a workflow with the tasks necessary to per-
form the query process and designate the participants
in the process, namely the data custodians. During
this phase, a governance board approval and other ad-
ministrative issues can also be included in the proto-
col. The next step is to use ATLAS to create a script
that defines the cohort and send it to the data custodi-
ans through the workflow execution.
After receiving the script, the DC runs it locally,
using a local installation of ATLAS, and generates
the results. Subsequently, the DC evaluates the re-
sults and decides if these can be shared or not. The
workflow management tool can be used to inform the
SM of the rejection and the respective reason. Oth-
erwise, the DC sends the results to the SM using the
same workflow management tool.
Once all data custodians have completed the local
queries and returned aggregate results, the SM uses
ATLAS to visualize the results and compiles them in
a document that is sent to the researcher, completing
the query process.
4 DISCUSSION
The worldwide proliferation of EHR systems leads to
the existence of an increasing number of digital clini-
cal data repositories. Despite the recognized value of
these repositories for secondary use, and their unde-
niable importance for clinical research, it is still very
difficult to access these data. There are several rea-
sons that make sharing of this data so difficult: the
existence of database silos, the difficulty in locating
EHR databases, the distribution and fragmentation of
the data, and privacy issues due to legal, ethical and
HEALTHINF 2018 - 11th International Conference on Health Informatics
130
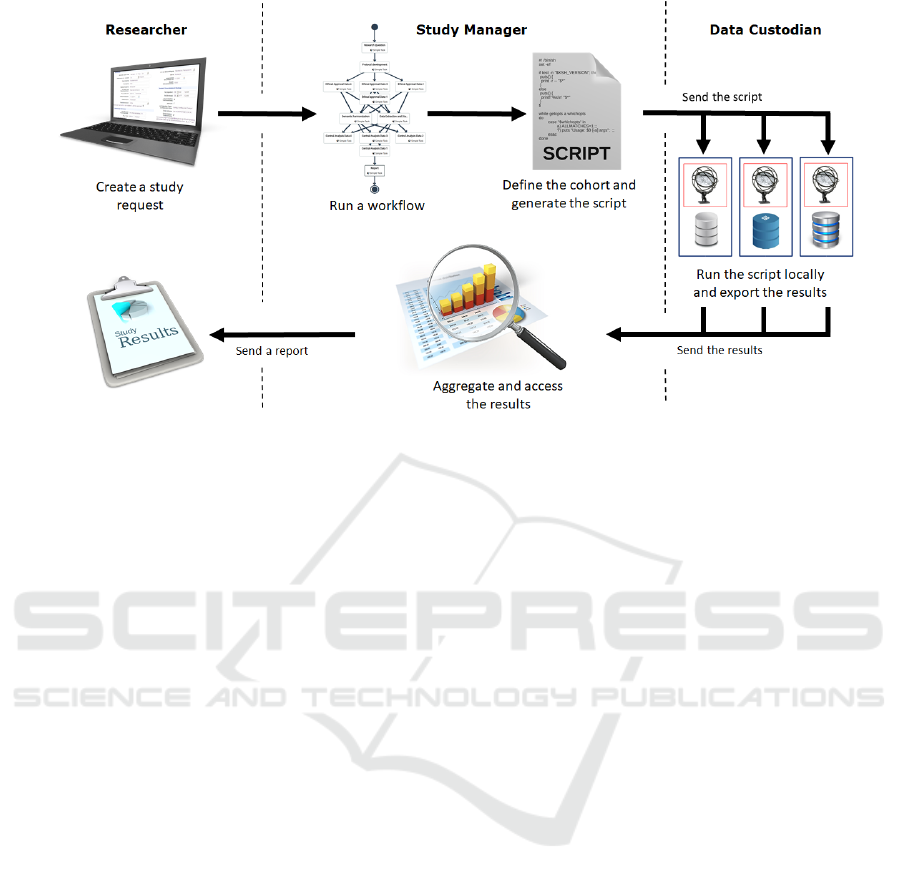
Figure 1: Workflow of the querying process.
regulatory requirements.
Technical solutions for health data integration typ-
ically use a centralized data warehouse, with replicas
of original EHRs, or a real-time distributed query sys-
tem, which relies on complex governance agreements
and institutions’ trust. These solutions are time con-
suming or imply governance models that might not be
allowed by most data custodians. Moreover, in both
cases, data custodians lose control of their data. Other
solutions are designed for a specific type of database
and are difficult to adapt to other types. Other solu-
tions suffer from technical complexity and most re-
search organizations do not have the technical skills
or knowledge to use them. However, there are par-
tial solutions, which can be combined. The EMIF
Data Catalogue, for example, enables researchers to
find several differentiated databases, and the OMOP
tools can transform data from different databases into
a CDM, thereby allowing queries across a set of
databases.
To overcome the continuing difficulties in the sec-
ondary use of EHRs, we developed a methodology
to perform semi-automatic distributed EHR database
queries. Our methodology does not use centralized
data warehouses, but rather it is semi-automatic so
that every data custodian can maintain control of their
database and only share the data they consider to
fulfill the legal, ethical and regulatory requirements.
Moreover, our methodology relies on existing solu-
tions and open-source software, which significantly
reduces the cost involved in the process.
Next, we present a simple example of a feasibil-
ity study involving the various actors (R, SM, and
DCs), i.e. a study to identify how many patients of
one or more databases fulfill some criteria. Here, we
ignore governance and contractual aspects, although
they can be incorporated at any stage of this work-
flow.
• Step 1 (R): Research question
– After logging into the EMIF Catalogue, the
user (researcher) fills out a form describing the
research question and the objective of the study.
– As an example, we may take a research ques-
tion such as “How many patients, with prostate
cancer, had prostate cancer screening” involv-
ing three databases. Other information, such
as the expected delivery deadline, the user’s
e-mail, institution and position, among others,
also need to be provided.
• Step 2 (SM): Feasibility assessment
– The study manager receives a notification about
the existence of a new study request.
– They log into the EMIF Catalogue and evaluate
the feasibility of this request.
– Through an internal messaging system, they
can ask the researcher for more information or
details, in order to better understand the scope
of the study. The study manager can also make
suggestions on how to improve the request.
– After this step is concluded, the study can start
internally.
• Step 3 (SM): Define the cohort template
– The study manager enters the ATLAS installa-
tion available in the EMIF Catalogue.
A Methodology to Perform Semi-automatic Distributed EHR Database Queries
131
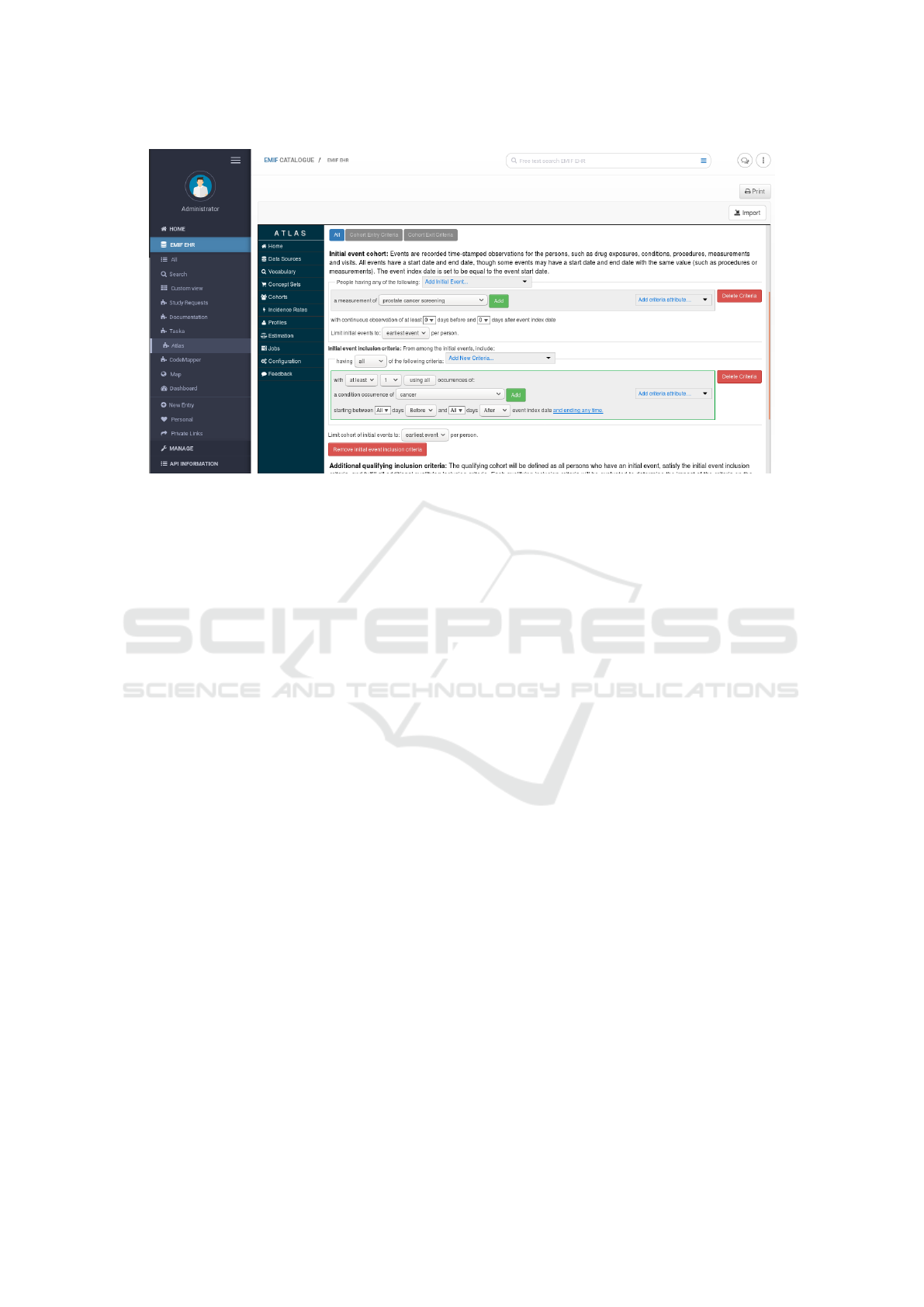
Figure 2: Example of a cohort definition.
– They start by creating the Concept Sets needed
for the Cohort definition, namely, “Prostate
cancer screening” and “Prostate cancer”.
– The Concept Sets created are used to specify
the inclusion criteria when the study manager
defines the cohort. Figure 2 presents the cohort
definition of this study.
– The cohort definition is exported in a JSON for-
mat.
• Step 4 (SM): Create and initiate the study work-
flow
– Using a workflow management tool, the study
manager creates a new study.
– They select the participants, namely, the data
custodians of the selected databases, assigning
them the tasks.
– After initiating the workflow, the study man-
ager shares the cohort definition, using the
workflow management tool.
• Step 5 (DC): Execute partial studies
– The study management tool sends a notifica-
tion to each data custodian selected by the study
manager, informing them that they have been
chosen as a participant in a study workflow and
have assigned tasks.
– The data custodian executes the common co-
hort definition using the local ATLAS installa-
tion.
– Results are analyzed locally, and evaluated re-
garding the possibility for sharing.
– If the results can be shared, the data custodian
exports them using the local ATLAS installa-
tion and uploads them into the study manage-
ment tool. Otherwise, the data custodian in-
forms the study manager that they will not share
the results.
• Step 6 (SM): Result integration and reporting
– After all data custodians complete their tasks,
the workflow management tool notifies the
study manager.
– They upload the file with the results sent by
each data custodian into the ATLAS installation
of the EMIF Catalogue.
– The study manager visualizes the results of
the study, using the ATLAS installation of the
EMIF Catalogue, and elaborates a report based
on these results.
– Through an internal messaging system, the
study manager sends the report back to the re-
searcher.
• Step 7 (R): Results evaluation
– The researcher receives a notification inform-
ing that the results are available.
– They access the results and analyse them.
– If needed, the researcher can ask for more infor-
mation, through the internal messaging system.
The presented methodology allows managing and
simplifying the execution of feasibility studies over
multiple EHRs databases, addressing one of the core
concerns for the sharing of clinical data for research,
i.e. by preserving local governance.
HEALTHINF 2018 - 11th International Conference on Health Informatics
132

5 CONCLUSIONS
In this paper we presented a methodology to perform
semi-automatic distributed EHR database queries that
uses preexisting partial solutions and open-source
software. The query process presented enables the
researcher to formulate a feasibility question and ob-
tain statistical and aggregated information about data
from different databases without accessing these data
directly or contacting the various data custodians.
ACKNOWLEDGEMENTS
This work has received support from the EU/EFPIA
Innovative Medicines Initiative Joint Undertaking
(EMIF grant n. 115372).
REFERENCES
Basti
˜
ao, S. L., D
´
ıas, C., van der Lei, J., and Oliveira, J. L.
(2014). Architecture to summarize patient-level data
across borders and countries. Studies in health tech-
nology and informatics, 216:687–690.
Cushman, R., Froomkin, A. M., Cava, A., Abril, P., and
Goodman, K. W. (2010). Ethical, legal and social
issues for personal health records and applications.
Journal of biomedical informatics, 43(5):S51–S55.
Daniel, C., Ouagne, D., Sadou, E., Forsberg, K.,
Mc Gilchrist, M., Zapletal, E., Paris, N., Hussain,
S., Jaulent, M.-C., and Kalra, D. (2016). Cross bor-
der semantic interoperability for clinical research: the
ehr4cr semantic resources and services. AMIA Sum-
mits on Translational Science Proceedings, 2016:51.
Doods, J., Botteri, F., Dugas, M., and Fritz, F. (2014).
A european inventory of common electronic health
record data elements for clinical trial feasibility. Tri-
als, 15(1):18.
Harris, S. B., Glazier, R. H., Tompkins, J. W., Wilton, A. S.,
Chevendra, V., Stewart, M. A., and Thind, A. (2010).
Investigating concordance in diabetes diagnosis be-
tween primary care charts (electronic medical records)
and health administrative data: a retrospective cohort
study. BMC health services research, 10(1):347.
Hersh, W. R. (2007). Adding value to the electronic health
record through secondary use of data for quality assur-
ance, research, and surveillance. Am J Manag Care,
81:126–128.
Hripcsak, G., Duke, J. D., Shah, N. H., Reich, C. G., Huser,
V., Schuemie, M. J., Suchard, M. A., Park, R. W.,
Wong, I. C. K., Rijnbeek, P. R., et al. (2015). Obser-
vational health data sciences and informatics (ohdsi):
opportunities for observational researchers. Studies in
health technology and informatics, 216:574.
Kahn, M. G., Batson, D., and Schilling, L. M. (2012).
Data model considerations for clinical effectiveness
researchers. Medical care, 50.
K
¨
opcke, F. and Prokosch, H.-U. (2014). Employing com-
puters for the recruitment into clinical trials: a com-
prehensive systematic review. Journal of medical In-
ternet research, 16(7).
Lopes, P., Silva, L. B., and Oliveira, J. L. (2015). Chal-
lenges and opportunities for exploring patient-level
data. BioMed research international, 2015.
Mann, C. (2003). Observational research methods. research
design ii: cohort, cross sectional, and case-control
studies. Emergency medicine journal, 20(1):54–60.
McDonald, H., Nitsch, D., Millett, E., Sinclair, A., and
Thomas, S. (2014). New estimates of the burden
of acute community-acquired infections among older
people with diabetes mellitus: a retrospective cohort
study using linked electronic health records. Diabetic
medicine, 31(5):606–614.
McMurry, A. J., Murphy, S. N., MacFadden, D., Weber, G.,
Simons, W. W., Orechia, J., Bickel, J., Wattanasin, N.,
Gilbert, C., Trevvett, P., et al. (2013). Shrine: en-
abling nationally scalable multi-site disease studies.
PloS one, 8(3):e55811.
Meystre, S., Lovis, C., B
¨
urkle, T., Tognola, G., Budrionis,
A., Lehmann, C., et al. (2017). Clinical data reuse
or secondary use: Current status and potential future
progress. IMIA Yearbook.
Miller, A. R. and Tucker, C. (2014). Health information
exchange, system size and information silos. Journal
of health economics, 33:28–42.
Murphy, S. N., Weber, G., Mendis, M., Gainer, V., Chueh,
H. C., Churchill, S., and Kohane, I. (2010). Serv-
ing the enterprise and beyond with informatics for
integrating biology and the bedside (i2b2). Jour-
nal of the American Medical Informatics Association,
17(2):124–130.
Ogunyemi, O. I., Meeker, D., Kim, H.-E., Ashish, N.,
Farzaneh, S., and Boxwala, A. (2013). Identifying
appropriate reference data models for comparative ef-
fectiveness research (cer) studies based on data from
clinical information systems. Medical care, 51:S45–
S52.
Ohmann, C. and Kuchinke, W. (2007). Meeting the chal-
lenges of patient recruitment. International Journal
of Pharmaceutical Medicine, 21(4):263–270.
Pakhomov, S., Weston, S. A., Jacobsen, S. J., Chute, C. G.,
Meverden, R., Roger, V. L., et al. (2007). Electronic
medical records for clinical research: application to
the identification of heart failure. Am J Manag Care,
13(6 Part 1):281–288.
Platt, R. and Carnahan, R. (2012). The us food and drug ad-
ministration’s mini-sentinel program. pharmacoepi-
demiology and drug safety, 21(S1):1–303.
Pringle, S. and Lippitt, A. (2009). Interoperability of elec-
tronic health records and personal health records: key
interoperability issues associated with information ex-
change. Journal of healthcare information manage-
ment: JHIM, 23(3):31–37.
Reisner, S. L., Vetters, R., Leclerc, M., Zaslow, S., Wol-
frum, S., Shumer, D., and Mimiaga, M. J. (2015).
Mental health of transgender youth in care at an ado-
lescent urban community health center: a matched ret-
A Methodology to Perform Semi-automatic Distributed EHR Database Queries
133

rospective cohort study. Journal of Adolescent Health,
56(3):274–279.
Ross, T. R., Ng, D., Brown, J. S., Pardee, R., Hornbrook,
M. C., Hart, G., and Steiner, J. F. (2014). The hmo
research network virtual data warehouse: a public data
model to support collaboration. EGEMS, 2(1).
Safran, C., Bloomrosen, M., Hammond, W. E., Labkoff, S.,
Markel-Fox, S., Tang, P. C., and Detmer, D. E. (2007).
Toward a national framework for the secondary use of
health data: an american medical informatics associ-
ation white paper. Journal of the American Medical
Informatics Association, 14(1):1–9.
Song, J. W. and Chung, K. C. (2010). Observational stud-
ies: cohort and case-control studies. Plastic and re-
constructive surgery, 126(6):2234.
Trifir
`
o, G., Coloma, P., Rijnbeek, P., Romio, S., Mosseveld,
B., Weibel, D., Bonhoeffer, J., Schuemie, M., Lei, J.,
and Sturkenboom, M. (2014). Combining multiple
healthcare databases for postmarketing drug and vac-
cine safety surveillance: why and how? Journal of
internal medicine, 275(6):551–561.
Weber, G. M., Murphy, S. N., McMurry, A. J., MacFad-
den, D., Nigrin, D. J., Churchill, S., and Kohane, I. S.
(2009). The shared health research information net-
work (shrine): a prototype federated query tool for
clinical data repositories. Journal of the American
Medical Informatics Association, 16(5):624–630.
HEALTHINF 2018 - 11th International Conference on Health Informatics
134
