
Sharing Genetic Data under US Privacy Laws
Michael Reep
1
, Bo Yu
1
, Duminda Wijesekera
1
and Paulo Costa
2
1
Department of Computer Science, George Mason University, Fairfax, VA, U.S.A.
2
Department of Systems Engineering and Operations Research, George Mason University, Fairfax, VA, U.S.A.
Keywords: Genetic Privacy, Electronic Medical Records, Ontology, Health Care, Genomic Medicine, SWRL (Semantic
Web Rule Language).
Abstract: Clinical medical practice and biomedical research utilize genetic information for specific purposes.
Irrespective of the purpose of obtaining genetic material, methodologies for protecting the privacy of
patients/donors in both clinical and research settings have not kept pace with rapid advances in genetics
research. When the usage of genetic information is not predicated on the latest laws and policies, the result
places all-important patient/donor privacy at risk. Some methodologies err on the side of overly stringent
policies that may inhibit research and open-ended diagnostic activity, whereas an opposite approach advocates
a high-degree of openness that can jeopardize patient privacy, inappropriately identify disease susceptibility
of patients and their genetic relatives, and thereby erode the doctor-patient privilege. As a solution, we present
a framework based on the premise that acceptable clinical treatment regimens are captured in workflows used
by caregivers and researchers and therefore their associated purpose are inherent to and therefore can be
extracted from these workflows. We combine these purposes with applicable consents that are derived from
applicable laws and practice standards to ascertain the releasability of genetic information. Given that federal,
state and institutional laws, rules and regulations govern the use, retention and sharing of genetic information,
we create a three-level rule hierarchy to apply the laws to a request and auto-generate consents prior to
releasing. Our hierarchy also identifies all pre-conditions that must be met prior to the genetic information
release, any restrictions and constraints to be enforced after release, and the penalties that may be assessed for
violating these terms. We prototype our system using open source tools, while ensuring that the results can
be added to existing Electronic Medical Records (EMR) systems.
1 INTRODUCTION
Genetic studies match genotypic and phenotypic data
to associate genetic markers with onset of diseases
(Ritchie et al., 2015). Multiple studies also show that
preventive care costs significantly less than treatment
upon disease onset and diagnosis (Németh et al.,
2013), (Pihoker et al., 2013). Furthermore, rapid
advancement of genetic research continues to
lengthen the list of predictable diseases. However,
both research and clinical use of genetic information
entail privacy challenges that differ from usage of
other medical data in following ways:
* Ethics: Privacy of genetic data differs from
traditional medical information privacy for example,
as protecting patients’ private information (e.g.,
Protected Health Information - PHI) is an ethical and
legal obligation. Data for genotype-phenotype
matching can be used to stigmatize or discriminate
against genetic relatives of a donor, so the dangers of
its exposure must be carefully weighed against the
benefits of its use (Ritchie et al., 2015), (Lowrance
and Collins, 2007), (McGuire and Gibbs, 2006).
There is an ongoing ethical debate between the two
different schools of thought, one in which the donor
gives open consent for using his/her data vs. the other
that advocates explicit purpose-based consent
(McGuire and Gibbs, 2006).
* Legal Issues: Due to the unusual situation of
being able to expose relative’s genetic composition,
genetic privacy has been proposed as categorical
privacy that differs from traditional individual-
centered concepts of privacy in literature (Lunshof et
al., 2008). Federal (HIPAA and GINA), state laws
and institutional polices provide the legal framework
for the sharing of genetic information. Furthermore,
genetic privacy laws vary from state-to-state and may
be inconsistent with, or more or less stringent than,
federal regulations.
Reep, M., Yu, B., Wijesekera, D. and Costa, P.
Sharing Genetic Data under US Privacy Laws.
DOI: 10.5220/0006550303490360
In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2018) - Volume 5: HEALTHINF, pages 349-360
ISBN: 978-989-758-281-3
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reser ved
349

* Social Implications: Societal views are often
reflected in law and/or organizational policies, so
their implications are likely inextricably intertwined
with laws and policies governing genetic privacy and
what constitutes informed consent.
As a solution, we provide an encompassing
framework consisting of workflow-enforced genetic
privacy as well as biomedical consent management,
consistent with state and federal genetic privacy laws
such as statute, regulation and precedent.
Following this Introduction, Section 2 addresses
related work; Section 3 reviews the prior work on the
prototype, Section 4 describes the overall architecture
and design for the implementation of our genetic
services workflow that enforces appropriate informed
consent based on applicable law to achieve genetic
privacy; Section 5 addresses the updates made to the
system as further state laws have been implemented,
Section 6 provides a specific example use Arizona
state laws, and, finally, Section 7 presents
conclusions.
2 RELATED WORK
Many researchers have suggested adopting traditional
information protection methodologies to protect
patients’ confidentiality. Yet, this might not be
effective due to the uniqueness of being traceable to
an individual or group of individuals (Mascalzoni et
al., 2008), (Gostin and Hodge, 1999). Some genetic
information of an individual may not only precisely
identify him/her as high risk of certain hereditary
disease(s), but also indicate that his/her relatives have
the same risks due to heritable genes.
Prince et al. describes three practical genetic
counselling cases that illustrate genetic
discrimination (Prince and Roche, 2014). The
fundamental covenant of protecting patient privacy is
embodied in patient-doctor privilege. Conversely,
many scholars believe that genetic information is
essentially familial in nature and is referred to as the
Genetic Information is Familial Thesis (GIFT) (Liao,
2009), because sharing such information will benefit
related groups of individuals. Some countries have
regulations to enforce sharing such information
among family members (Lucassen and Kaye, 2006),
(ASHG, 1998). However, many publications discuss
and debate the familial approach, with their authors
advocating the view that humans possess the rights of
privacy and to protect those that do not want to know
(Liao, 2009), (ASHG, 1998). Conversely, rapid
innovations in genetic research require wide
accessibility to many genetic databases. The idea of
open access in the field of genomic research is
expressed in the Bermuda Principles and the Fort
Lauderdale Agreement, which has been applied in
North America and in the UK for funded research
(Sherlock, 2009). Genetic research typically requires
additional metadata with genetic data sets, such as
demographic details family relationships, medical
history, etc. These metadata elements can be
exploited for tracing an individual’s identity.
In general medicine, an informed consent,
especially informed privacy consent, provides the
proper opportunity and knowledge for patients and
research participants to understand and decide how
the medical community can use and share their
identifiable medical information, in addition to the
risks and benefits of treatment regimes. Analogously,
informed consent tailored for genetic research,
clinical usage and counselling constitutes a strong
basis for ensuring appropriate genetic privacy. Some
genetic medical practices and biomedical research are
performed without obtaining appropriate informed
consent such as enticing participants in a study
without obtaining the proper informed consent. To
address this issue, some researchers advocate
different methodologies such as using highly-
stringent policies to maintain patient confidentiality,
but this approach potentially risks limiting scientific
innovation (Kaye et al., 2012). Yet, other researchers
have proposed a new, open-consent model for
medical and scientific genetic research (Lunshof et
al., 2008) or open-access policies for genetic data
sharing (Hallinan and Friedewald, 2015).
EMRs play a vital role of sharing medical
information among participating actors based on their
usage scenarios. Using EMRs for genetic services
present a unique set of challenges (Kaye et al., 2009).
Belmont et al. highlighted the privacy, ethical and
legal issues of handling genetic data in EMRs
(Mascalzoni et al., 2008). The study conducted by
Scheuner et al. to validate if current EMR systems
meet genetic information needs (Belmont and
McGuire, 2009) shows an overall lack of support for
functionality, structure, and tools for clinical genetic
practice. A more recent study of the state of EMRs
supporting genomics for personalized medicine
identifies structure of data as a challenge (Scheuner et
al., 2009).
As a solution, an approach based on the premise
that acceptable clinical treatment regimens are
captured in workflows used by caregivers and
researchers and therefore their associated purpose are
inherent to and therefore can be extracted from these
workflows (Reep et al., 2016).
HEALTHINF 2018 - 11th International Conference on Health Informatics
350
Some researchers suggested that the legislation
for generating and using genetic information properly
is pivotal to improving genetic privacy (Ullman-
Cullere and Mathew, 2011). In 2013, the Health
Insurance Portability and Accountability Act of 1996
(HIPAA) Omnibus Rule included genetic
information as PHI to be regulated under the privacy
portion of HIPAA. Nonetheless, states may have
different definition of genetic information. The
combination of Federal privacy laws along with the
various state laws form a fragmented regulatory and
statutory landscape for permissible information
sharing and consent management. To be valid,
informed consents for genetic privacy must comply
with these laws and regulations. Indeed, significant
regulatory gaps create additional burdens in
providing automated ways to obtain and generate
information consent in EMRs.
3 PRIOR WORK
Releasing genetic medical information involves
addressing a number of unique considerations not
present for other types of medical records. Genetic
information is a component of protected health
information where the individual’s identity may be
embedded directly into the data structures. In
addition, the genetic information provides insight into
almost every aspect of an individual’s health. Within
the United States, the special characteristics have
resulted in laws, regulations and policies targeting the
criteria where genetic information can be released,
the requirements (or preconditions) that must be
fulfilled before information release, and obligations
that must be enforced once the information has been
released. We previously proposed a mechanism to
address the problem space using three distinct
components:
▪ Workflow to gather information, execute a
rules engine, display the outcome, obtain
acceptance from the user of the results, and
enforce requirements associated with
information release.
▪ An ontological rule-base that takes the data
from the workflow, evaluates the applicable
laws, determines the pre-conditions and
obligations, and decides on the releasability of
genetic data.
▪ A consent service that interacts with the
workflow engine and the ontology to pass data
back and forth. The service includes the Rule
Hierarchy Algorithm which combines the
outcomes from the three levels (Federal, State
and Organization) and provides a final result
for permitting or denying access.
We have expanded the number of states that are
incorporated in the prototype. In order to address the
wide range of situations reflected in these laws, we
have implemented a number of changes to the
ontology, workflow and rules to process the actual
States laws. The major changes have been in the
workflow component and are addressed in the rest of
this paper along with other corresponding
modifications to the ontology and consent service.
4 ARCHITECURE AND DESIGN
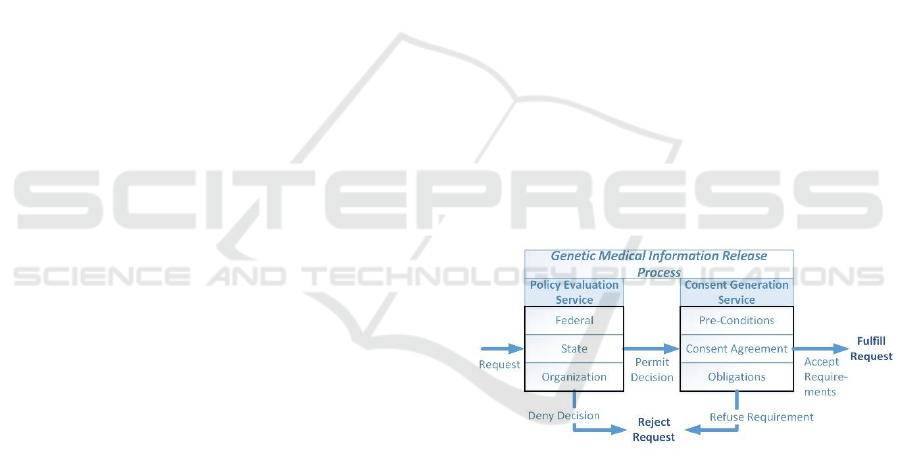
The process to release genetic medical information is
based on two related set of processes as seen in Figure
1. In the first process for policy evaluation, each set
of laws, rules and regulations at the Federal, State and
medical organization levels are examined for
applicability to the request being made. The request
may be either to perform medical procedures used to
obtain genetic test results or for the genetic
information from the tests contained in the medical
record. The outcome of the Policy Evaluation Service
will either allow the request to continue, potentially
based on enforcing specific consent requirements, or
deny the request outright.
Figure 1: Release Processes.
One of the main components of the policy
evaluation is to generate the requirements under
which the requested access can be granted. These
requirements encompass one or more activities
related to verifying that that any consent agreements
have been signed, indicated pre-conditions are met,
and that the enforcement mechanism for post-release
obligations have been established. The activities are
combined into a consolidated set for enforcement if
there are multiple rules that meet the evaluation
criteria.
Sharing Genetic Data under US Privacy Laws
351
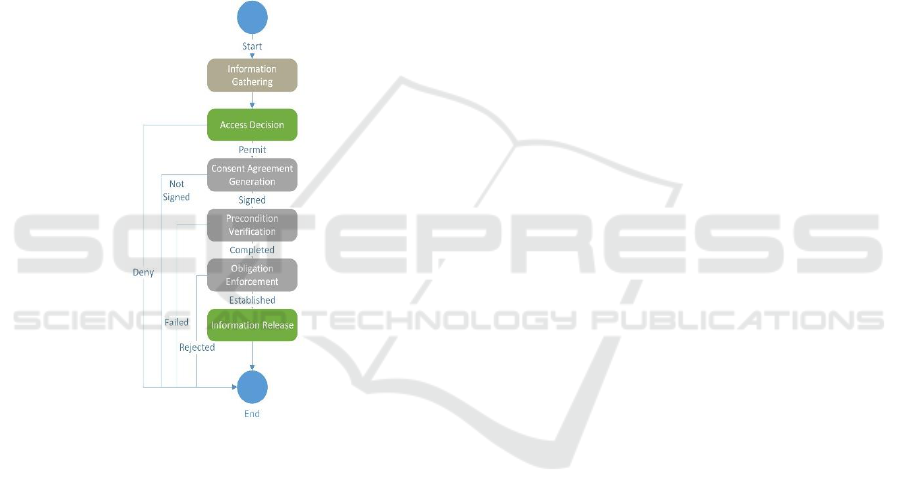
4.1 System Architecture Abstraction
The system workflow to enforce consent
requirements orchestrates the various components
necessary to invoke genetic information protection as
shown in Figure 2. After the information on the
subject, request and the requester is gathered, the
access decision is made by the policy evaluator. If a
permit decision is returned, the consent generation
service uses the workflow engine to display the
individual pre-conditions for validation along with
generating the text for the required consents, and
enforcing the obligations associated with the data
release. The information released is performed once
the workflows steps for the consent generation
process is completed. .
Figure 2: Workflow Construct.
4.2 Policy Design
Our policies are written as a collection of rules that
use three main abstract entities with their associated
attributes:
▪ Request: this abstraction incorporates the
subject of the request, the purpose for requesting the
information, and the resource (part of the medical
record) that the request addresses.
▪ Requester: the person/entity making the request
to access the medical information including their role,
their organization, and other auxiliary attributes of
this organization.
▪ Response: the determination applying the
appropriate rules to come to an access decision The
Response includes a list of any pre-conditions to be
verified before the information is released, specific
consent clauses that the subject must sign, and
obligations that must be enforced upon information
release. A separate Response instance is created for
each of the Federal, State and Organization levels that
are then evaluated for a final decision.
Our policies consist of rules that codify the policy.
The Purpose, Resource and State (where the request
is being made) are required as the minimum data set
with other components added to address specific
situations. For example, a request to access data for
the Law Enforcement purpose will require either the
Requester’s Role or Organization relationships to
perform the validations. If the rule generates a Permit
Access response, then any requirements (pre-
conditions, consent clauses and obligations) are
dictated and used to populate the Response.
The Federal and State rules include an option for
an “override” capability so that the other levels can
change the permission already established. For
example, if the Federal rule grants access but the State
laws are allowed to deny the access request, the
Federal response override flag is set to true. The
override flags are used in conjunction with Federal,
State and Organization responses to generate a
consolidated Final Access Decision.
Each rule is structured to identify the criteria
under which it is applicable, the outcome of whether
access is permitted or denied, and any requirements
placed on an information release. The rule criteria
includes:
▪ Purpose the information will be used for once
released (Required)
▪ Request Target as either a specific test to be
performed or genetic medical information (Required)
▪ State in which the request is made (Required)
▪ Requester attributes such as whether parent or
guardian (Optional)
▪ Requester role such as in law enforcement
(Optional)
▪ Requester’s organization such as associated with
medical facility (Optional)
▪ Subject attributes such as whether of consent age
(Optional)
The output of the rule sets the following
properties:
▪ Level that generated the rule (Required as
Federal, State or Organization)
▪ Access Permission is granted (Required as
Permit or Deny)
▪ Override Allowed for the rule at lower level
(Required for Federal or State levels as true or false)
▪ Decision Source to trace back specific text
generating the rule (Required)
▪ Pre-conditions that must be validated prior to
HEALTHINF 2018 - 11th International Conference on Health Informatics
352

release (Optional and may be more than one)
▪ Consent Clauses that must be accepted (Optional
and may be more than one)
▪ Obligations that must be enforced upon
information release (Optional and may be more than
one)
▪ Penalties if specified pre-conditions or
obligations are not met (Optional and may be more
than one).
4.3 Policy Evaluator Design
The Policy Evaluator uses the rules in order to
generate an access decision and, when appropriate,
the associated pre-conditions, consent text and
obligations that are associated with the genetic
information release. The workflow gathers the
information used in the rule evaluation either through
querying the user or accessing external data sources
such as the facility EMR. The Policy Evaluator is
described in Algorithm 1.
Algorithm 1: EvaluatePolicies.
Input: Workflows, RuleBase
FOR EACH entity (Request, Requester) (1)
READ data values from workflow (2)
POPULATE current entity properties (3)
FOR EACH rule (4)
EVALUATE rule criteria (5)
IF rule criteria met (6)
RETRIEVE associated Response Instance
(Federal, State, Organization) (7)
SET Response Decision for rule (8)
IF Decision = “Permit” (9)
ADD Preconditions, Consent
Clauses, Obligations to
Response (10)
IF Precondition OR Obligation
HAS Penalties (11)
ADD Penalties to Response (12)
END IF
END EACH
SET Final Response = Federal Response (13)
If State Access Decision = Final Access Decision
AND Final Access Decision =
“Permitted” (14)
ADD State Response Preconditions,
Consent Clauses, Obligations and
Penalties to Final Response (15)
IF State Access Decision <> Final Access Decision
AND Federal Override = TRUE (16)
SET Final Access Decision = State Access
Decision (17)
IF State Access Decision = “Permitted” (18)
ADD State Response Preconditions,
Consent Clauses, Obligations
and Penalties to Final
Response (19)
SET Final Override = State Override (20)
IF Organization Access Decision = Final Access Decision
AND Final Access Decision =
“Permitted” (21)
ADD Organization Response Preconditions,
Consent Clauses, Obligations and
Penalties to Final Response (22)
IF Organization Access Decision <> Final Access
Decision AND
Final Override = TRUE (23)
SET Final Access Decision = Organization Access
Decision (24)
IF Organization Access Decision =
“Permitted” (25)
ADD Organization Response,
Preconditions, Consent Clauses,
Obligations and Penalties to
Final Response (26)
RETURN Final Decision and related Preconditions,
Consent Clauses, Obligations and Penalties (27)
At a high-level the Policy Evaluator process is as
follows:
1. Retrieve request and requester information
gathered from workflow and populate the process
entities. (1-3)
2. Execute each rule that is applicable to the Request
and Requester properties. (4-5)
3. If the rule is applicable, store the output to the
corresponding response entity including pre-
conditions, consent text, obligations for an
information release and penalties for failing to
enforce requirements for access decisions. . (6-12)
4. Use the Rules Hierarchy Algorithm to combine the
Federal, State and Organization outcomes and
determine the final result (permit or deny) along with
assembling the preconditions, consent clauses,
obligations and penalties (13-26)
5. Return the final results components so that the
Consent Generation Process can be performed (27).
4.4 Consent Generation Service Design
The Consent Generation Service processes the policy
evaluator output when a Final Decision is made to
potentially permit the disclosure of genetic
information. First, the service enforces all
requirements set by the policies prior to allowing the
genetic information release. As seen below, the
consent agreement signature is obtained, every pre-
condition validated, and all obligations set in order for
the information release to the requester. Once the
algorithm is completed and the release decision is set,
the information is passed back to the workflow for
display to the requester. If the releases is approved,
Sharing Genetic Data under US Privacy Laws
353
the EMR can then provide the genetic information to
the requester.
The high-level algorithm for the Consent
Generation Service is described in Algorithm 2.
Algorithm 2: GenerateConsent.
Input: Workflows, Final Decision
SET release = TRUE
IF Final Decision includes Consent Clauses (1)
CREATE Agreement (2)
FOR EACH Consent Clause (3)
ADD Consent Clause TO
Agreement (4)
OBTAIN Signature on Agreement (5)
IF signature NOT Obtained (6)
SET release = FALSE (7)
END IF (8)
IF Final Decision includes Pre-Conditions (9)
FOR EACH Pre-condition (10)
VALIDATE Pre-Condition Met (11)
IF Pre-Condition NOT Met (12)
SET release = FALSE (13)
END EACH (14)
END IF (15)
IF Final Decision INCLUDES Obligations (16)
FOR EACH Obligation (17)
SET Obligation enforcement (18)
IF Obligation NOT Enforced (19)
SET release = FALSE (20)
END EACH (21)
END IF (22)
RETURN release (23)
At a high-level the Consent Generation process is
as follows:
1. Initialize release flag to be true (1)
2. If there are any consent clauses associated with
the information release, create a new agreement and
then add all the consent clauses from each rule into
one document for the subject’s signature (2-5). Deny
the release if no signature is obtained (6-7).
3. If there are any pre-conditions associated with
the information release, validate that each one has
been successfully met (9-11). Deny the release if any
pre-condition is not validated (12-13).
4. If there are any obligations associated with
the information release, set the enforcement
mechanism for each one (16-18). Deny the release if
any obligation is not set for enforcement (19-20).
5. Return the resulting release value to the
workflow and EMR.
5 IMPLEMENTATION
ENHANCEMENTS
This section describes how we prototyped our model
as described in our previous paper and expanded the
prototype with new functionality to include
refinements as we have implemented laws from
additional states. These improvements are the focus
of the rest of this paper.
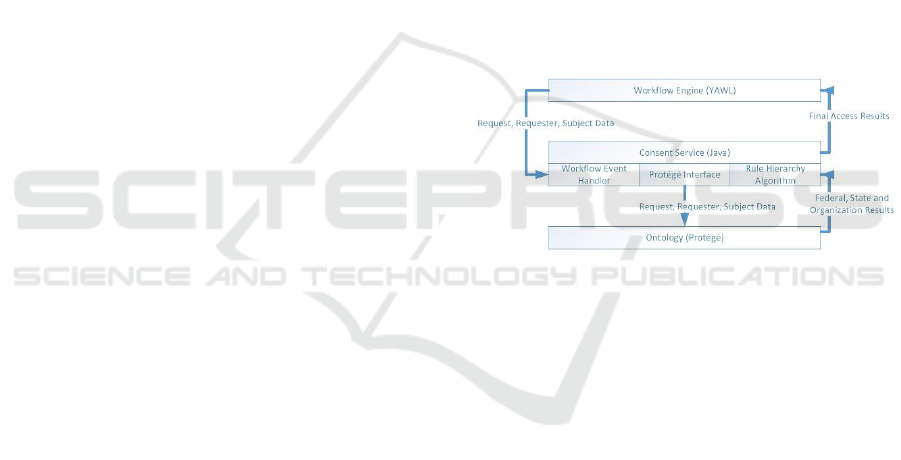
Figure 3 shows the interactions between the
workflow engine, the Consent Service and the
ontology. The combination of these components
implements the Policy Evaluation Service and the
Consent Generation Service to provide privacy
protection for genetic medical information. The
workflow component is implemented using YAWL
(Yet Another Workflow Language). The ontology
and associated rules for policy evaluation was
developed with Protégé and the DL Reasoner. The
consent service was developed in Java for the
interactions between the ontology and workflow. In
addition, the Rules Hierarchy Algorithm was
implemented using Java due to the limitation of DL
addressing specific negation situations inherent in
laws and policies.
Figure 3: Prototype Components.
5.1 Workflow Map Upgrade
The primary focus on our recent research efforts has
been on enhancing the workflow component to better
reflect the consent process needed to obtain
permission to release data by collecting all pre-
conditions and then implementing the associated
obligations for post-release. The updates improve the
process for releasing genetic information and
ensuring privacy protections by separating out the
pre-condition activities prior to information release
approval and the obligations enforcement required
after the information release.
HEALTHINF 2018 - 11th International Conference on Health Informatics
354
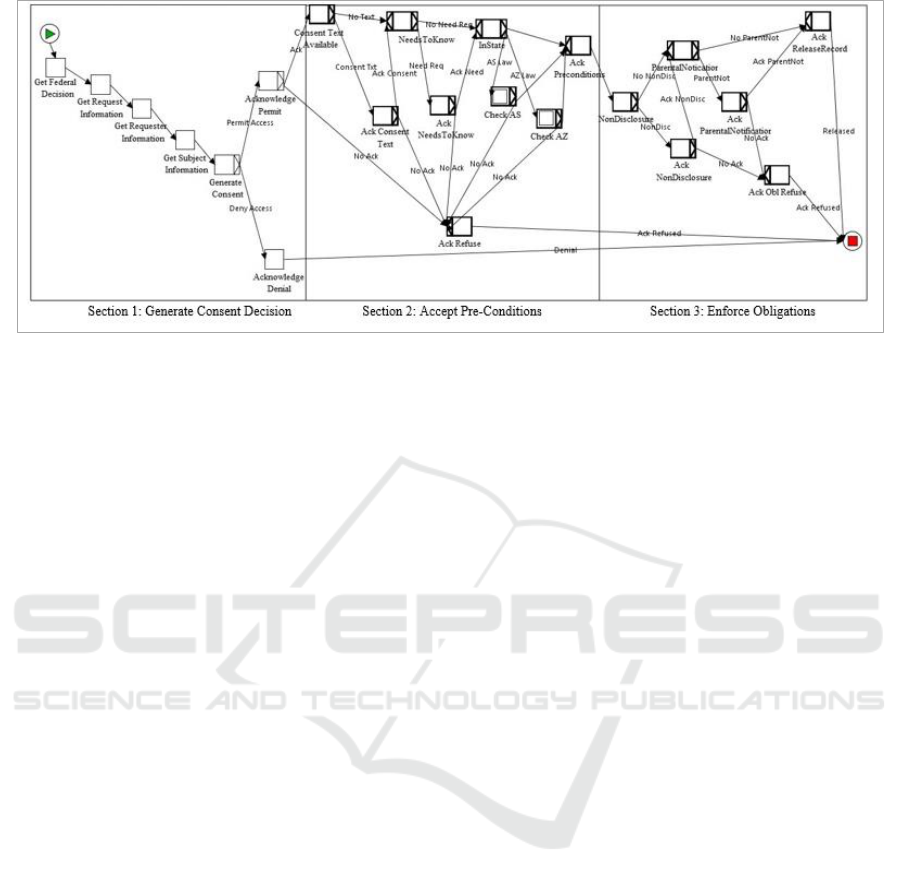
Figure 4: Genetic Privacy Workflow.
As shown in Figure 4, there are three major
sections to the workflow which is implemented in
YAWL. In the first section, the consent workflow
performs the Information Gathering regarding the
permissibility of access in relation to Federal laws
along with data properties for the request, the
requester and the subject. The “Generate Consent”
step uses an event handler in the workflow that is tied
to Java code in the Consent Service. As described
below, the Consent Service makes the Access
Decision on whether access will be permitted for the
user along with collating all the pre-conditions,
obligations and consent text from each level (Federal,
State and Organization). These are used by the
workflow for the user to acknowledge the results
If consent is granted, then in Section 2 the
“Consent Text Available” step implements the
Consent Agreement Generation to display the
specific language for all consent clauses so the
subject can electronically sign the consent agreement.
The Pre-Condition Verification is performed and the
user must acknowledge that each individual
precondition is met with a separate confirmation for
each one. During this section, the state-specific pre-
conditions are also checked in sub workflows via the
“InState” gate.
Once all the pre-conditions have been
acknowledged, the workflow moves into Section 3 to
establish the Obligation Enforcement mechanisms for
any obligations that must be enforced with the
permission. If the user fails to acknowledge any of the
pre-conditions or obligations, the workflow states that
situation to the user and permission is denied. At the
end of the workflow, the results are returned to the
associated EMR to perform the actual information
release if approved.
5.2 Implementing Policies using
Ontologies
The ontology changes introduced since the previous
prototype encompass both additional attributes to
capture specific conditions along with several
structure changes as seen in Figure 5. (Changes from
the previous ontology model are in italics.) The
structural changes were as follows:
▪ Adding an Activity class to support State laws
for obtaining consent prior to performing activities
related to genetic privacy such as genetic testing. (As
opposed to requests for Resources which is contained
in the medical record generated after the activity was
completed.
▪ Separating out requirements that must be
enforced prior to information release (Precondition
class) from those that must be enforced after the data
is released for use (obligations).
▪ Adding a Penalty class to articulate the possible
outcomes should the pre-condition or obligation
requirements are not met.
In addition to these structural changes, additional
Boolean data properties were added to Purpose,
Subject, Requester, Role and Organization classes in
order to support conditions associated with specific
rule processing.
For example, the Subject class in Figure 5
provides selected information about the person who
is the patient or client in the medical records being
accessed for genetic information. A Boolean flag was
added to Subject (isDeceased) to address a genetic
information release under Arizona law (AZ 12-
2802.E) for when the subject is deceased. Since other
Arizona law (AZ 12-2802.A.6) permitted information
release when the health care provider (physician or
organization) ordered a genetic test (attribute) or if
Sharing Genetic Data under US Privacy Laws
355
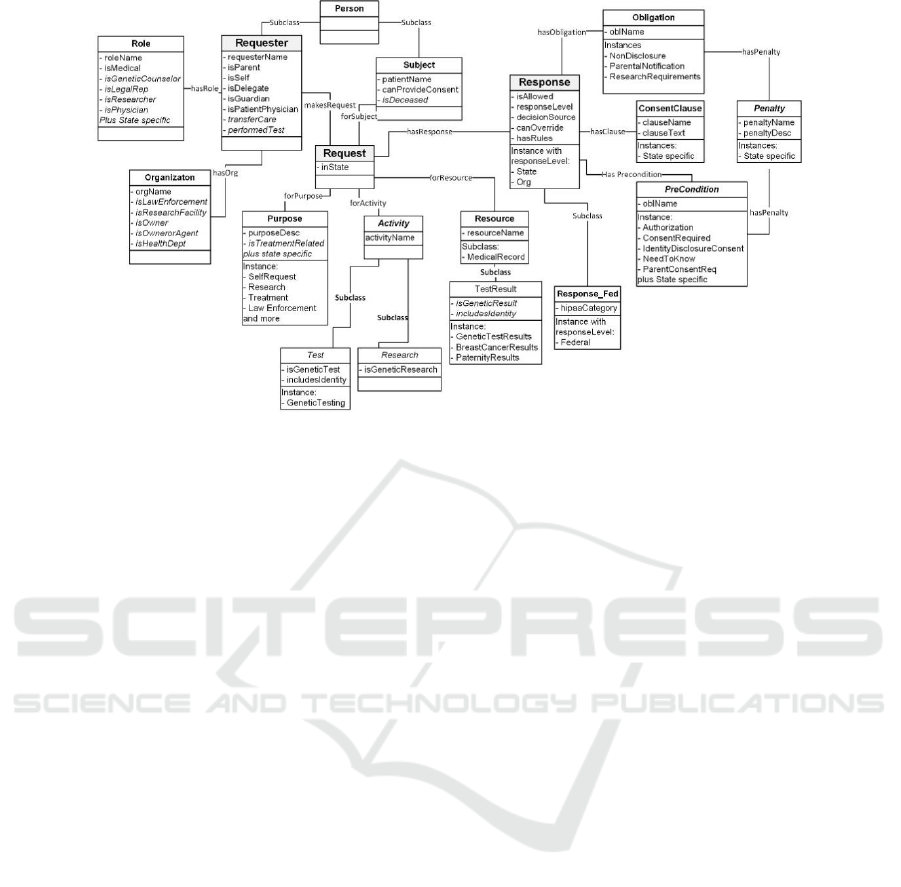
Figure 5: Genetic Privacy Ontology.
care was transferred from a provider that had access
to genetic information (attribute) (AZ 12-2802.A.11),
the Requester data properties now includes
performedTest and transferCare flags. Similar
situations required additions to the other classes.
More flags are expected as additional laws,
regulations and policies are added to the ontology
Another set of Boolean data properties were
added to abstract out specific aspects of dealing with
genetic data. In the first case, because some genetic
tests only deal with specific parts of the genome that
do not identify a specific individual (such as for a
specific disease), an includeIdentity flag is used to
provide additional restrictions when the test includes
protected information like those used in law
enforcement. In addition, Boolean data properties
were added to permit enforcement of the genetic
restrictions without listing individual tests or test
results. The properties are isGeneticTest (Test
subclass), isGeneticResearch (Research subclass)
and isGeneticResult (TestResult subclass). The
ontology contains only the information from the
EMR that is necessary to implement genetic privacy
rules.
5.3 Automatically Generating Consents
The Consent Service serves as the integration engine
between the workflow/EMR and the ontology. Once
the request, requester and subject attributes are
gathered in the workflow steps (Section 1 of the
workflow) and used to populate the workflow
variables, the Consent Service is triggered
by the workflow engine, as the next step, that is
GenerateConsent.
The service collates the data from the workflow
variables for subject, request and requester, populates
the ontology, invokes the rules processor, retrieves
the intermediate results from the ontology, invokes
the Rule Hierarchy Algorithm to reconcile the
Federal, state and organization level results, and
finally generates the final access permissions. The
final permissions are transferred back to the workflow
along with the associated pre-conditions and
obligations. The outcome includes the consolidated
list of conditions for all three levels. For example, the
list of consent clauses required by both the Federal
regulations and organizational policies.
Our initial prototype was modelled on preliminary
work associated with representative state laws. As
described above, we have identified specific
attributes that are needed to implement new scenarios
as we have implemented the full set of laws from
additional states. So far the primary difference to the
consent service from our initial prototype involves
supporting the ontology changes for additional
classes and properties in transferring data between the
ontology and workflow. In addition, these class and
property changes impacted the Rules Hierarchy
Algorithm with the additions of the PreCondition and
Penalty classes.
In the Rules Hierarchy Algorithm, the Result
variables for the Answer, Pre-conditions,
Obligations, Decision Source, Clauses, Penalties, and
Rules are initialized to the corresponding federal
variables, which were retrieved from Protégé. The
Federal Override variable is then evaluated to
determine if other rules are to be evaluated. If so, the
HEALTHINF 2018 - 11th International Conference on Health Informatics
356

algorithm checks for existing State answers and, if
found, determines if the Federal and State answer
match. The system adds the State variables to the
Result variables when the Federal and State match
while the Results variables are set to the State results
when there is no match.
For the Organization level, the algorithm
determines if there is an Organization result and if
there is a State result with a State Override flag set to
true or there is no State answer. If the Results are the
same, then the Algorithm adds the Organization
variables to the Result variables otherwise the Results
variables to be equal the Organization values if results
are different and the override flag is set to true. At
the end of processing the Results variables are passed
back to the workflow engine.
6 ARIZONA CASE STUDY
As a case study, Arizona permits access to genetic
information for purposes not explicitly stated in the
law if consent is obtained first. (AZ 12-2802.A.2).
The first YAWL screen shown in Figure 6 is for
the Get Request Information step in the workflow
process to describe why the request is needed, what
part of the medical record is to be accessed, in what
state the action is being performed and an option to
get permission to perform an activity (such as Genetic
Testing) instead of accessing the genetic information
resource. The Get Requester Information shows the
key meta-data needed for the requester and the related
entities such as organization and role. Each of the four
Get steps have a similar screen. (Because Federal
laws are well established and addressed in current
policies, our focus at this time is on the
implementation of state laws. Therefore the Federal
access permission and override option is gathered
using a graphical user interface.)
The ontology is populated with data from the
workflow after all the information gathering steps
are completed. The ontological rules engine is
invoked and the rule specific for this case study are
executed. The SWLR rule for this condition is:
Rule: makesRequest(?r, ?req), inState(?req, "AZ"),
forResource(?req, ?resource),
isGeneticResult(?resource, true), forPurpose(?req,
?pur), isAZAllowed(?pur, false), hasResponse(?req,
?resst), responseLevel(?resst, "State"),
oblName(?pre, "ConsentRequired"),
clauseName(?clause, "AZGeneticAuthorization")
Figure 6: Workflow Entry Screen Shots.
isAllowed(?resst, true), canOverride(?resst, false),
decisionSource(?resst, "AZ LAW 12-2802.A"),
hasPreCondition(?resst, ?pre), hasClause(?resst,
?clause), hasRule(?resst, 57)
In this rule,
▪ ?r is for the Requester of the Request
▪ ?req is for the Request that links the various
components, such as Subject, Purpose and Resource
▪ ?pur is the Purpose that is associated with the
Request
▪ ?resst is the State Response object that is
associated with the Request.
▪ ?resource is for the “GeneticTestResults” part of
the medical record
▪ ?pre has the pre-condition that ConsentRequired
must be obtained for this rule
▪ ?clause indicates the consent agreement for the
patient must include the AZGeneticAuthorization
clause
These SWRL statements are explained in Table I.
Sharing Genetic Data under US Privacy Laws
357

Table 1: Sample Pre-Condition Rule.
SWRL Statement
Explanation
makesRequest(?r, ?req)
Links Requester for the
Request
inState(?req, "AZ")
Request is for Arizona
forResource(?req,
?resource)
Links Request with the
Resource
isGeneticResult(?resource,
true
Restricts the rule to a
Resource that is identified as
a genetic test results
forPurpose(?req, ?pur)
Links Request with Purpose
isAZAllowed(?pur, false)
Restricts the rule to only
execute when the purposes is
not specifically allowed in
Arizona
hasResponse(?req, ?resst)
Links the Request with a
Response to store answer
responseLevel(?resst,
"State")
Gets the Response for State
level to store answers
oblName(?pre,
"ConsentRequired")
Gets the Pre-Condition with
the name for Consent
Required
clauseName(?clause,
"AZGeneticAuthorization")
Gets the Clause for Arizona
authorization
-> isAllowed(?resst, true)
Sets the State response to
access is allowed
canOverride(?resst, false)
Sets the state Response to
not allow override by
organization
decisionSource(?resst,"AZ
LAW 12-2802.A")
Sets the State response to
reflect the decision source as
state law
hasPreCondition (?resst,
?pre)
Links the retrieved Pre-
Condition with the State
response
hasClause(?resst, ?clause)
Links the retrieved Clause
with the State response
hasRule(?resst, 57)
Sets the rule number to 57
for reference
When the Pellet reasoner finds the instance for
access in Arizona for a genetic test result based on a
purpose not specifically addressed, the rule is
executed and the ?resst data properties are populated
with the indicated values. In addition, ?pre and
?clause instances are associated with the response as
conditions to accessing the record. (The rule links the
permission to access the genetic information with any
associated pre-conditions, obligations and consent
clauses.)
The reasoner output for the AZ State result is
shown in Figure 7. The output also includes
references to a second rule and the NonDisclosure
obligation. In Arizona, genetic information releases
also require the enforcement of a non-disclosure
requirements (AZ 12-2802.F) which is reflected in
Rule 54. As the last steps for the Consent Service
processing, the service extracts the response
information from the ontology. The results are then
evaluated using the Rule Hierarchy Algorithm to
combine the responses for the Federal, State and
Organizational rules into the final decision.
Figure 7: AZ Response.
Upon completion of the Consent Service
invocation, the results are passed back to the
workflow. The “AcknowPermit” screen in Fig 8
shows the results for granting access permission
displayed for validation by the user. This screen
shows the outcome to the user from the ontology rule
processing and the Rules Hierarchy Algorithm
evaluation.
Once the user acknowledges the overall results in
Figure 9, the workflow then ensures that each pre-
condition is completed prior to genetic information
release. Each pre-condition clause is evaluated for
applicability in this case and the appropriate actions
taken to enforce the requirement. The individual pre-
conditions are displayed and accepted separately to
develop an audit trail of acceptance and to ensure all
requirements are acknowledged.
In the AZ case study, the workflow first displays
the consent text and requires that the clauses be
accepted by the subject for the information release as
seen in Figure 9. (The YAWL screen will be replaced
with a digital signature implementation upon
integration with an EMR system.)
Once the generic pre-conditions that are
applicable to all states have been evaluated, the main
workflow in Figure 4 goes to the ”In State” step to
determine if there are additional pre-conditions based
on the state where the information request is being
performed. This attribute-based determination
evaluation is used to reduce unnecessary steps in the
workflow.
HEALTHINF 2018 - 11th International Conference on Health Informatics
358
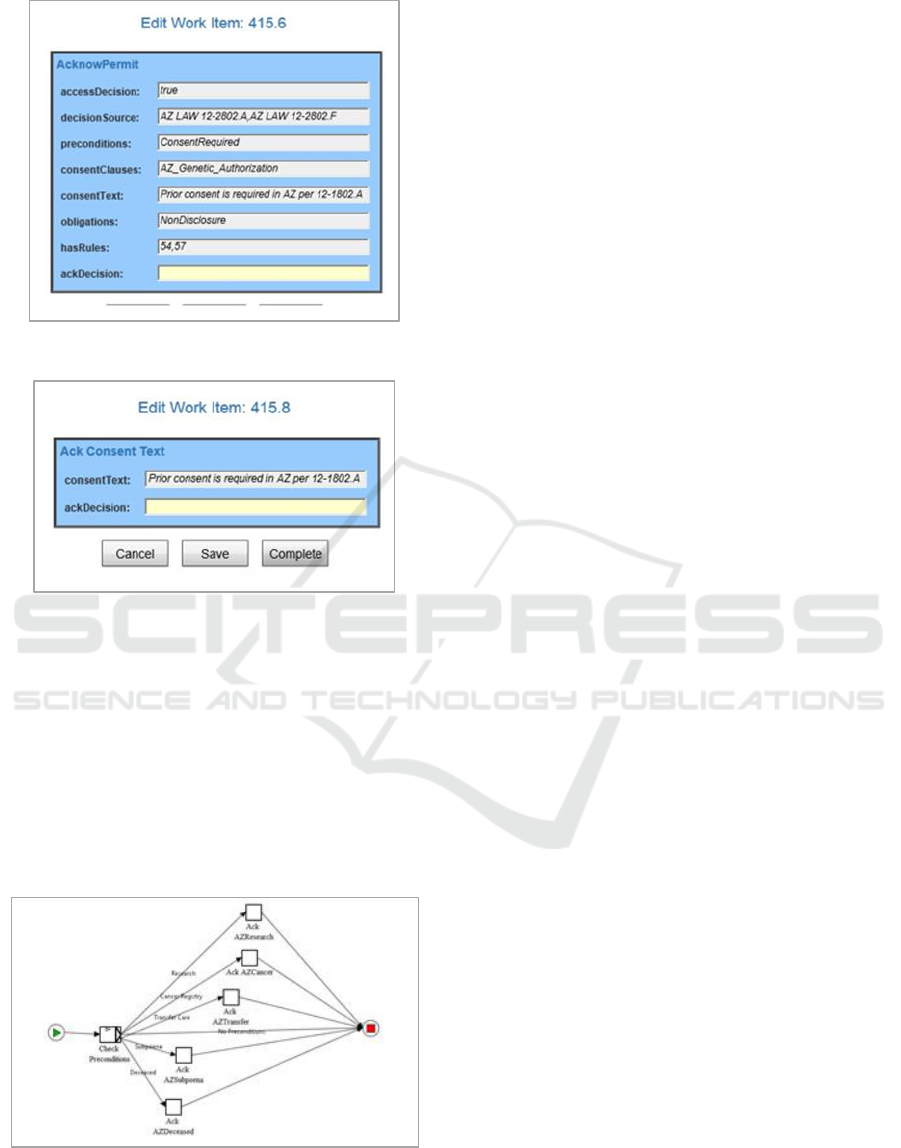
Figure 8: AZ YAWL Results Confirmation.
Figure 9: AZ YAWL Results Confirmation.
Separate sub-workflows then enforce the
requirements for that state through user validation for
each specific requirement. The AZ sub-workflow is
shown in Figure 10 which has separate requirements
to address situations for genetic research, the state
cancer registry, transferring care between providers,
information release under subpoenas, and deceased
subjects. Any of the conditions would generate a
separate confirmation screen to ensure the applicable
pre-conditions have been met.
Figure 10: Arizona Sub-Workflow.
Failing to complete any pre-conditions moves the
workflow to the Ack Refuse step as seen in Figure 4
and then to the subsequent end of the workflow
without permitting information release. If all the pre-
conditions are met, the workflow moves to enforcing
the obligations associated with the actions as seen in
Section 3 of the workflow diagram in Figure 5. Each
obligation also has a separate acknowledgement to
ensure the appropriate actions are taken.
For this case study, an additional confirmation
screen is displayed for the NonDisclosure obligation
seen in Figure 8. (The state enforces a requirement
that all genetic results may not be disclosed beyond
the person or organization that receives the
information.) Upon completion of the obligation
steps, the workflow ends and the information release
of genetic information occurs with all federal, state
and local laws, rules, and regulations implemented
and enforced.
7 CONCLUSIONS
We provide a framework to ensure the appropriate
availability of genetic medical information while
enforcing the privacy protections. The expanded
prototype works to bring together the applicable
operational data in an EMR workflow into our
framework to provide a definitive and consolidated
response for access and the associated pre-
conditions/obligations for information disclosure.
While we continue to implement additional Federal
and State rules to develop a comprehensive repository
and rule base, our ongoing work focus on the
interactions with representative policies and
procedures for a medical organization. The pre-
conditions and obligations will undergo further
analysis to formalize the interactions and pro-actively
identify potential conflicts within the rule set. This
intersection will allow rules to be generated based on
the risk of releasing protected privacy information.
We expect the resulting prototype to demonstrate the
overall capabilities needed to meet the medical
community’s access requirements while balancing
the individual rights to privacy and ownership of their
genetic medical data.
REFERENCES
Ritchie, M., Holzinger, E., Li, R., Pendergrass, S. and Kim,
D. (2015). "Methods of integrating data to uncover
genotype-phenotype interactions." Nature Reviews
Genetics 16.2. 85-97.
Németh, A., Kwasniewska, A., Lise, S., Schnekenberg, R.,
Becker, E., Bera, K. and Talbot. K. (2013) "Next
Sharing Genetic Data under US Privacy Laws
359

generation sequencing for molecular diagnosis of
neurological disorders using ataxias as a model."
Brain. awt236.
Pihoker, C., Gilliam, L., Ellard, S., Dabelea, D., Davis, C.,
Dolan, L. and Mayer-Davis, E. (2013). "Prevalence,
characteristics and clinical diagnosis of maturity onset
diabetes of the young due to mutations in HNF1A,
HNF4A, and glucokinase: results from the SEARCH
for Diabetes in Youth." The Journal of Clinical
Endocrinology & Metabolism 98.10. 4055-4062.
Lowrance, W. and Collins, F. (2007). “Identifiability in
genomic research.” SCIENCE 317. 600-602.
McGuire, A. and Gibbs, R. (2006). "No longer de-
identified." SCIENCE-NEW YORK THEN
WASHINGTON- 312.5772. 2006. 370.
D’Abramo, F., Schildmann, J. and Vollmann, J. (2015).
"Research participants’ perceptions and views on
consent for biobank research: a review of empirical
data and ethical analysis." BMC medical ethics 16.1.
2015. 1.
Lunshof, J., Chadwick, R., Vorhaus, D. and Church, G.
(2008). "From genetic privacy to open consent." Nature
Reviews Genetics 9.5. 2008. 406-411.
The Health Insurance Portability and Accountability Act of
1996 (HIPAA). Pub. L. 104-191, 110 Stat. 1936,
codified as amended at 42 U.S.C x300gg and 29 U.S.C
x1181 et seq. and 42 U.S.C x1320d et seq.
Genetic Information Non-discrimination Act of 2008
(GINA). Pub. L. 110-233, 122 Stat. 883, codified as
amended in scattered sections of 26, 29, and 42 U.S.C.
Mascalzoni, D., Hicks, A., Pramstaller, P. and Wjst, M.
(2008). "Informed consent in the genomics era." PLoS
Med 5.9. e192.
Gostin, L., and Hodge Jr., J. (1999) "Genetic privacy and
the law: an end to genetics exceptionalism."
Jurimetrics. 21-58.
Prince, A. and Roche, M. (2014). "Genetic information,
non-discrimination, and privacy protections in genetic
counseling practice". Journal of genetic counseling
23.6. 891-902.
Liao, S. (2009) "Is there a duty to share genetic
information?". Journal of medical ethics 35.5. 306-309.
Lucassen, A and Kaye, J. (2006). "Genetic testing without
consent: the implications of the new Human Tissue Act
2004." Journal of medical ethics 32.12. 690-692.
American Society of Human Genetics Social Issues
Subcommittee on Familial Disclosure, ASHG
STATEMENT Professional Disclosure of Familial
Genetic Information. 1998) Am. J. Hum. Genet. 62:
474–483.
Sherlock, E. (2009) “Disclosure of patient's genetic
information without their consent- Is the "public
interset" really a Sufficient Justification?”. Genomics
Law Report. 2009. Available at:
http://www.genomicslawreport.com/index.php/2009/1
1/10/disclosure-of-patientsgenetic-information-
without-their-consent-is-the-public-interest-really-a-
sufficient-justification/ . [Accessed 2 March 2015].
Kaye, J., Gibbons, S., Heeney, C., Parker, M. and Smart,
A. (2012). "Governing biobanks: Understanding the
interplay between law and practice." Bloomsbury
Publishing.
[Hallinan, D. and Friedewald, M. (2015). "Open consent,
biobanking and data protection law: can open consent
be ‘informed’under the forthcoming data protection
regulation?" Life sciences, society and policy 11.1. 1.
Kaye, J., Heeney, C., Hawkins, N., De Vries, J. and
Boddington, P. (2009). "Data sharing in genomics—re-
shaping scientific practice". Nature Reviews Genetics
10.5. 331-335.
Mascalzoni, D., Hicks, A., Pramstaller, P. and Wjst, M.
(2008). "Informed consent in the genomics era." PLoS
Med 5.9. e192.
Belmont, J. and McGuire, A. (2009). "The futility of
genomic counseling: essential role of electronic health
records." Genome medicine 1.5. 1.
Scheuner, M., de Vries, H., Kim, B., Meili, R., Olmstead,
S. and Teleki, S. (2009) "Are electronic health records
ready for genomic medicine?". Genetics in Medicine
11.7. 510-517.
Ullman-Cullere, M. and Mathew, J. (2011) "Emerging
landscape of genomics in the electronic health record
for personalized medicine." Human mutation 32.5. 512-
516.
Gymrek, M., McGuire, A., Golan, D., Halperin, E. and
Erlich, Y. (2013) "Identifying personal genomes by
surname inference." Science 339.6117. 2013. 321-3.
Reep
,
M., Yu, B., Wijesekera
,
D. and Costa, P. (2016),
“Sharing Data under Genetic Privacy Laws”. In:
Proceedings of the Eleventh Conference on Semantic
Technology for Intelligence, Defense, and Security.
[online] Fairfax: CEUR Workshop Proceedings, pp. 46-
54. Available at: http://ceur-ws.org/Vol-1788/
[Accessed: 25 July 2017].
HEALTHINF 2018 - 11th International Conference on Health Informatics
360
