
Multiple Sclerosis Lesion Segmentation using Improved Convolutional
Neural Networks
Erol Kazancli
1,2
, Vesna Prchkovska
3
, Paulo Rodrigues
3
, Pablo Villoslada
4
and Laura Igual
1
1
Department of Mathematics and Computer Science, Universitat de Barcelona,
Gran Via de les Corts Catalanes, 585, 08007 Barcelona, Spain
2
DAMA-UPC, Universitat Polit
`
ecnica de Catalunya, C. Jordi Girona, 1-3, 08034 Barcelona, Spain
3
Mint Labs Inc. 241 A Street Suite 300, 02210 Boston, MA, U.S.A.
4
Center of Neuroimmunology, Institut d’Investigacions Biomediques August Pi Sunyer (IDIBAPS),
Villarroel, 170, 08036 Barcelona, Spain
Keywords:
Multiple Sclerosis Lesion Segmentation, Deep Learning, Convolutional Neural Networks.
Abstract:
The Multiple Sclerosis (MS) lesion segmentation is critical for the diagnosis, treatment and follow-up of
the MS patients. Nowadays, the MS lesion segmentation in Magnetic Resonance Image (MRI) is a time-
consuming manual process carried out by medical experts, which is subject to intra- and inter- expert variabi-
lity. Machine learning methods including Deep Learning has been applied to this problem, obtaining solutions
that outperformed other conventional automatic methods. Deep Learning methods have especially turned out
to be promising, attaining human expert performance levels. Our aim is to develop a fully automatic met-
hod that will help experts in their task and reduce the necessary time and effort in the process. In this paper,
we propose a new approach based on Convolutional Neural Networks (CNN) to the MS lesion segmentation
problem. We study different CNN approaches and compare their segmentation performance. We obtain an
average dice score of 57.5% and a true positive rate of 59.7% for a real dataset of 59 patients with a specific
CNN approach, outperforming the other CNN approaches and a commonly used automatic tool for MS lesion
segmentation.
1 INTRODUCTION
Multiple Sclerosis (MS) is a chronic neurological di-
sease that afflicts especially the young population be-
tween the ages 20 and 50. It affects 2.3 million pe-
ople worldwide and can cause symptoms such as loss
of vision, loss of balance, fatigue, memory and con-
centration problems, among others. It remains a very
challenging disease to diagnose and treat, due to its
variability in its clinical expression (National MS So-
ciety, 2017). MS is characterized by lesions throug-
hout the brain that are caused by the loss of mye-
lin sheath around neurons in the brain, which is also
known as demyelination. The lesions are visible in se-
veral modalities of Magnetic Resonance Image (MRI)
with different contrasts. The number and the total vo-
lume of MS lesions are indicative of the disease stage
and are used to track disease progression.
The accurate segmentation of lesions in MRI is
important for the accurate diagnosis, adequate treat-
ment development and patient follow-up of the MS
disease. Manual segmentation of MS lesions by ex-
perts is the most commonly used technique and is still
considered to produce the most accurate results alt-
hough it suffers from many complications. First of
all, it is subject to intra- and inter- expert variability,
which means there are significant differences between
two segmentations performed by two different experts
(due to slightly varying definitions) or by the same
expert at different times (due to fatigue or similar fac-
tors). Secondly there is a shortage of adequately trai-
ned experts given the huge amount of segmentation
need. Thirdly, the segmentation task requires valuable
expert time and concentration, which could ideally be
dedicated to other tasks. These drawbacks make it
necessary and desirable to develop a semi-automatic
segmentation method that would assist experts in the
task with a reduced amount of time and intra- inter-
expert variability or, in the ideal case, a fully auto-
matic segmentation method which would obviate the
need for experts and produce accurate/reproducible
results.
260
Kazancli, E., Prchkovska, V., Rodrigues, P., Villoslada, P. and Igual, L.
Multiple Sclerosis Lesion Segmentation using Improved Convolutional Neural Networks.
DOI: 10.5220/0006540902600269
In Proceedings of the 13th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2018) - Volume 4: VISAPP, pages
260-269
ISBN: 978-989-758-290-5
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

Several methods previously presented in the lite-
rature resort to machine learning approaches. Some
methods use supervised approaches with hand-crafted
features or learned representations and some other
methods use unsupervised approaches like clustering
which aim to detect lesion voxels as outliers. Ex-
amples of supervised models used in MS segmenta-
tion tasks are k-nearest neighbour methods, artificial
neural networks, random decision forests and baye-
sian frameworks among others (Garcia-Lorenzo et al.,
2013). Examples of unsupervised models are fuzzy c-
means or Gaussian mixture models with expectation
maximization (EM) (Garcia-Lorenzo et al., 2013).
Unsupervised models suffer from non-uniformity in
the image intensities and lesion intensities since this
variability cannot be captured by a single global mo-
del (Havaei, 2016). In this respect supervised met-
hods present an advantage, potentially being able to
capture this variability with the appropriate choice of
training set or features.
Recently, Deep Learning (DL) has been very
successful in the Computer Vision area, achieving
improvements in accuracies sometimes as high as
30% (Plis et al., 2014). The main strength in DL,
also differentiating it from other machine learning
methods, is its automatic feature extraction capability.
Normally, raw data has to be processed automatically
or manually to extract meaningful and useful featu-
res through a process commonly known as ”feature
engineering”. This process requires time and careful
analysis, and includes subjectivity, which might bias
the results or produce erroneous results. However,
in DL, the feature extraction is data-driven using an
appropriate loss function and learning algorithm for
Deep Neural Networks, which removes the subjecti-
vity, randomness and expert knowledge to a certain
degree. Moreover, the features obtained are hierarchi-
cal, each network layer producing more abstract fea-
tures using the less abstract features obtained in the
previous layer. Thus feature extraction is carried out
step-by- step, which is likelier to produce more com-
plex and useful features. Another strength of DL is
its ability to represent very complex functions, which
might also be considered as its drawback since it is
prone to easily over-fit. However, the over-fitting can
be prevented with the correct guidance and regulariza-
tion methods. DL methods are also robust to outliers,
which is very common in neuroimaging data (Good-
fellow et al., 2016), (Bengio, 2012) and (Deep Lear-
ning, 2017).
Previous work on MS lesion segmentation with
DL is generally developed using voxelwise classifica-
tion (lesion vs. normal) and is done on 2D/3D patches
centered on the voxel of interest to obtain a complete
segmentation of the whole brain (Greenspan et al.,
2016). There are also some studies considering the
whole image as input and performing a segmentation
in a single step as in (Brosch et al., 2015) and (Brosch
et al., 2016). In some methods global context is pro-
vided to the network, in addition to the local con-
text, to give more information about the nature of a
voxel (Ghafoorian et al., 2017). Convolutional Neu-
ral Networks (CNNs) are commonly used as part of
the architecture due to their strong feature extraction
capabilities dealing with images (Vaidya et al., 2015),
while Restricted Boltzmann Machines (RBMs) and
Auto-encoders are generally exploited to obtain a
good initialization of the network, which might affect
the ultimate performance, as shown in (Brosch et al.,
2015) and (Brosch et al., 2016).
In this paper, we propose a MS lesion segmen-
tation method based on a voxelwise classification on
MRI with DL using a combination of different appro-
aches presented in the literature together with our own
contributions. We explore a new sub-sampling met-
hod to improve the learning process and develop a
new Convolutional Neural Networks (CNN) approach
to achieve better performance results. The aim of this
study is to achieve a method that will surpass the per-
formance of existing methods in helping the experts
in the MS lesion segmentation work and even make
their interruption minimal. We compare different ap-
proaches with DL so far applied to MS Segmentation.
2 METHODOLOGY
In this section, we present our strategies for data
pre-processing, sub-sampling of the training set, de-
signing the CNN architecture and developing diffe-
rent approaches to improve the segmentation perfor-
mance.
2.1 Data Pre-processing
We have, at our disposal, T1 and T2 MRI modali-
ties, tissue segmentation and manual lesion segmenta-
tions of 59 subjects from Hospital Cl
´
ınic (Barcelona).
The tissue segmentation was performed by Freesurfer
v.5.3.0 toolbox (Freesurfer, 2013). The manual seg-
mentation was performed by an expert / two experts
from Hospital Cl
´
ınic team. The voxel resolution of
the MRIs is 0.86mm x 0.86mm x 0.86mm and the
image size is 208 x 256 x 256. As a pre-processing of
the MRI images we apply skull stripping, bias-field
correction, tissue-segmentation and co-registration.
Additionally we apply 0-mean unit-variance normali-
Multiple Sclerosis Lesion Segmentation using Improved Convolutional Neural Networks
261

zation to the data. For this process we use the Scikit-
Learn’s preprocessing package (Scikit-Learn, 2010).
2.2 Sub-sampling Strategy
We pose the segmentation problem as a voxelwise
classification throughout the MRI image; therefore,
by a sample we mean a 3D patch centered on the voxel
of interest (to be classified). Initially we consider pa-
tches of size 11x11x11. A positive sample is such
a patch centered on a lesion voxel, while a negative
sample is a patch centered on a non-lesion voxel.
The data comes with a big imbalance of positive-
negative samples; negative samples greatly outnum-
bering the positive ones, because the lesion regions
generally make up a very small proportion of the
whole brain. To overcome this problem in the trai-
ning set, we take all the positive samples and select
as many negative samples using two different appro-
aches: random sampling and sampling around the le-
sions.
By random sampling we mean choosing negative
samples randomly throughout the brain, without ta-
king into account its location. By sampling around
lesions we mean taking negative samples very close
to the lesion areas. The latter approach produced bet-
ter results in the initial experiments therefore we kept
to this approach in the final experiments. This might
be due to the fact that in the random sampling met-
hod, the selected negative samples are very similar to
each other and does not represent the diversity of the
negative samples. However, when we select the nega-
tive samples around the lesions, we add more variety
and harder cases to the training set. In addition, we
avoid the negative examples very close to (2-voxels)
lesion regions, since these voxels may be in reality le-
sion voxels although they were not labeled as such by
the expert.
Finally, it is important to note that we select the
samples from white and gray matter although the stu-
dies so far generally chose their data from only white
matter since the probability to have a lesion in the
white matter is far greater than having it in the gray
matter.
2.3 Patch-based Classification using
CNN
Our initial CNN architecture is based on the
study (Vaidya et al., 2015) with several differences.
We use a simple CNN architecture with two convolu-
tional layers and one fully connected layer. The pa-
tches are 3D cubes centered on the voxel of interest,
which are obtained from T1 and T2 images. The first
convolutional layer uses 60 kernels of size 4x4x4 with
a stride value of 1 and no padding. This layer is fol-
lowed by an average pooling layer that takes 2x2x2
patches of the output generated in the previous layer
and produces an average for each patch. The stride
value for average pooling is 2. The second convoluti-
onal layer consists of 60 kernels of size 3x3x3 with
a stride value of 1 and no padding. After this se-
cond layer of convolutions we use average pooling of
2x2x2 patches with a stride value of 2 and followed
by a fully connected layer. This layer consists of 200
hidden units that are fully connected to the output of
the previous layer.
In the study (Vaidya et al., 2015) authors use 3 mo-
dalities T1, T2, and FLAIR, but we use only two, T1
and T2 since we do not have the FLAIR modality for
our patients. The patch size they use is 19x19x19 but
since our images have nearly half the resolution of the
images used in the study we choose our initial patch
size as 11x11x11. They consider different modalities
as input channels and start from 4D data. We chose to
start with separate branches from different modalities
and merge them at the fully-connected layer since our
experiments show improvements with this approach.
Also our subsampling method which was explained in
the previous section, differs from theirs.
For the convolutional layers and fully connected
layer ReLU (Rectified Linear Unit) is used as acti-
vation function. The output layer is a softmax layer
of two units, which produces a probability for the two
classes, which are lesion and non-lesion for the center
voxel. The class with a higher probability is the deci-
sion class produced. To compute gradients and guide
the training of the network cross-entropy loss function
is used. We use mini-batch gradient descent with a
batch size of 128 for each training step. To adjust
the convergence speed of the algorithm, an adaptive
learning method, namely Adam (Adaptive Moment
Estimation) is used which adapts the momentum and
learning rate throughout the training. At the convolu-
tional layers, batch normalization is applied, which is
known to lead to faster convergence and which serves
as a type of regularization method. We apply dropout
to the fully connected layer to add further regulariza-
tion. The number of epochs is set experimentally. An
scheme of the proposed architecture can be seen in
Figure 1.
2.4 Improvements of the Original
Model
Starting from the first model described above, we test
several approaches with different experimental set-
tings and improvements of the original approach: le-
VISAPP 2018 - International Conference on Computer Vision Theory and Applications
262
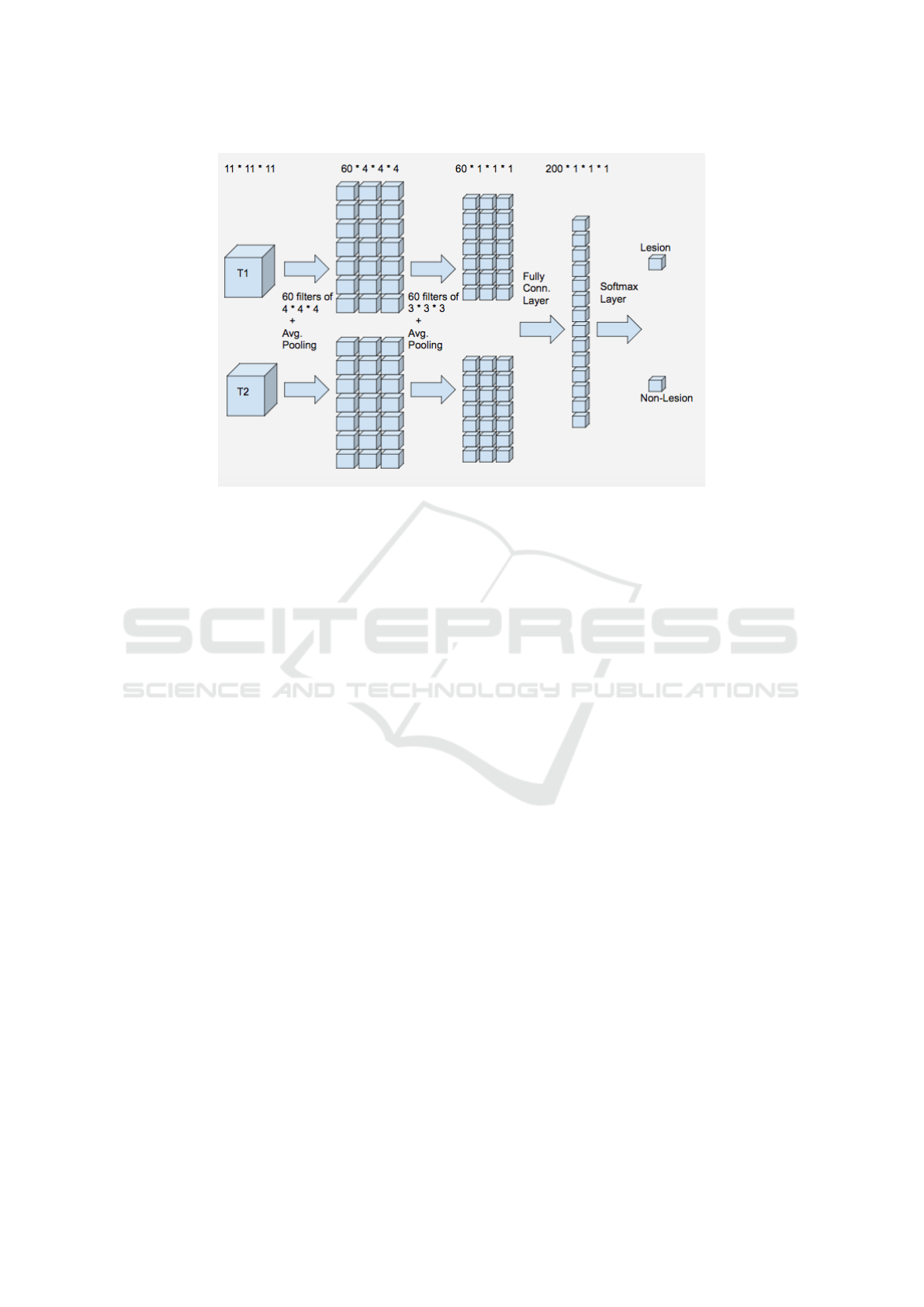
Figure 1: Proposed CNN Architecture.
sion location information, multi-class classification
and cascade approach.
Lesion Location Information. As a second appro-
ach, we add the lesion location information to the fe-
ature set to test if it improves the results. We think
this information might be useful since the probability
of having a lesion in certain regions of the brain might
show differences. For this approach we store the x, y,
z coordinates of each patch, we normalize them and
add these features at the fully connected layer. The
additional computational cost by adding these 3 fea-
tures is negligible for training and testing.
Patch Size. As the third approach, we increase the
patch size from 11x11x11 to 19x19x19 to see if gi-
ving more information of the context improves the
results. Note that increasing the patch-size increases
the number of initial features cubically. This means
increasing the computational cost substantially. We
keep the location information for this approach and
further approaches.
Cascade. Up to this point, the main problem in the
obtained results of our approaches is the high num-
ber of false positives obtained. Even though true ne-
gative rates reached levels of 96% the resulting seg-
mentation contained a high number of false positives,
even surpassing the number of true lesion voxels, due
to the high number of non-lesion voxels in the brain
compared to the lesion voxels. This is undesirable for
an automatic segmentation technique since the human
expert will need to discard these false positives, which
might even make the automatic segmentation useless.
The next approach focuses on decreasing these false
positives.
In the fourth approach, we implement two CNNs
in a cascade fashion as proposed in the study of (Val-
verde et al., 2017). We implement a CNN as explai-
ned before (in the second approach, with 11x11x11
patch size and location information) and obtain a first
model. With this model, we segment all the training
subjects automatically, which are used in the selection
of training samples for the second stage. For the se-
cond model, we use exactly the same architecture as
in the first stage but the sampling method differs. For
positive samples, we choose all the lesion voxels; For
the negative samples, we choose as many non-lesion
voxels in such a fashion that half of this number co-
mes from the false positives, which were wrongly
classified negative samples in the first stage, and the
other half comes from the true negatives from the first
stage segmentation results. The reason to choose the
false positives is to be able to remove these false po-
sitives in the second stage and the reason to choose
from true negatives is to prevent the model to forget
the knowledge obtained in the first model. In the ori-
ginal study negative samples are only selected from
the false negatives, which in our case performed far
from desired. The second stage model is thus trained
with the samples selected as just explained. In the tes-
ting stage, an initial segmentation is obtained using
the first CNN model. The candidate lesions obtained
from the first model are fed into the second model and
a final segmentation is obtained with the resulting po-
sitives of the second model. The computational cost
to the training of this model is more than twice the
cost of a one-stage model, since it also includes the
evaluation of all the training samples. However, once
Multiple Sclerosis Lesion Segmentation using Improved Convolutional Neural Networks
263
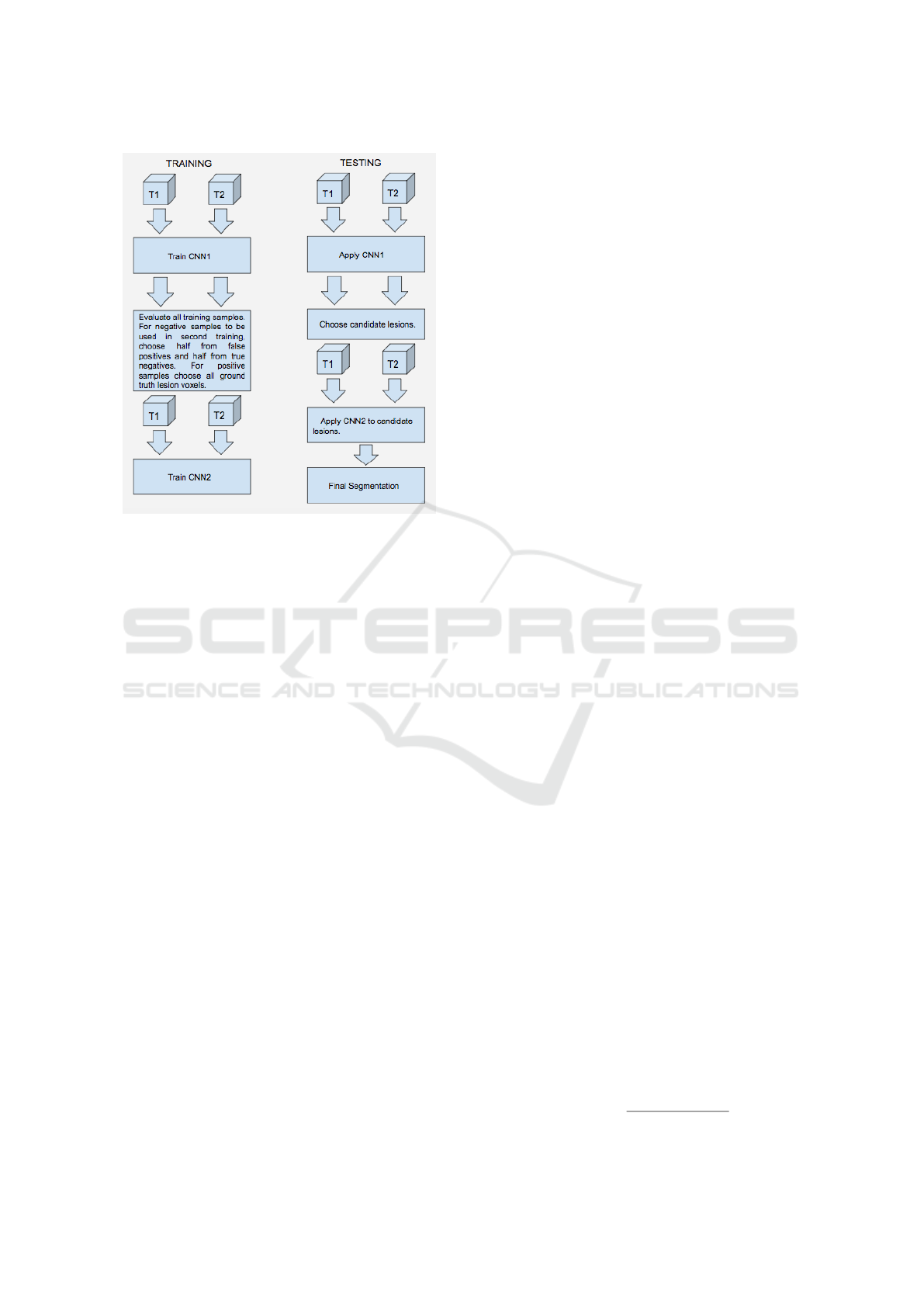
Figure 2: Two stage training and testing - Cascade.
the training is obtained the computational cost for tes-
ting does not double since the samples evaluated in
the second stage are a very small proportion of the
whole sample set. Figure 2 illustrates the cascade ap-
proach.
With the cascade implementation we should re-
duce the number of false positives at the cost of losing
also some of the true positives, but hopefully the gain
is considerably higher than the loss.
Multi-class Classification Problem. We realize that
there is a difference in the accuracy rates between the
region along the border of a lesion and the region far-
from a lesion border. This observation brought to our
minds to try first a 4-class model and subsequently
a 3-class model. For the fifth approach, we incre-
ase the number of classes from 2 (lesion, non-lesion)
to 4 (lesion interior, lesion border, non-lesion border,
non-lesion interior) during training to try to reduce
the false positive rate. During testing, we merge the 4
classes back to 2 classes.
The problem with the 4-class model is that the
number of lesion interior voxels is quite low com-
pared to the number of other classes and to balance
the training set we have to decrease the number of
samples substantially. This is something undesirable,
thus we consider lesion voxels as one class, and di-
vide the non-lesion voxels into two classes, which are
border non-lesion, interior non-lesion. Thus, for the
sixth approach, we consider 3 classes (lesion, border
non-lesion, interior non-lesion).
As a last step, we test the cascade implementation
with this 3-class model as a seventh approach. This is
the exact same cascade approach explained before in
the fourth approach, but with 3 classes instead of 2.
2.5 Technical Specifications
In order to develop our DL approaches we use an EC2
instance of type p2.xlarge of Amazon Web Services.
This is a cloud service of Amazon with GPU that pro-
vides high computational power for computationally
intensive processes such as DL (Amazon, 2017). It
also comes with an execution environment that con-
tains DL frameworks such as Tensorflow, Caffe, The-
ano, Torch, etc. We use Python as a programming
language and Tensorflow as a DL framework to im-
plement and run our DL algorithms. We also use
cloud storage provided by Amazon to store our trai-
ning samples. We handle the neuroimaging files in
the NIfTI-1 format with the Nibabel library of py-
thon (Nibabel, 2017).
3 EXPERIMENTS AND RESULTS
In this section, we describe the validation strategy
(data distribution and validation measures), the expe-
riments and obtained results.
3.1 Validation Strategy
As we already described in section 2.1, the ground-
truth (GT) is made by T1 and T2 MRI modalities,
tissue segmentation and manual lesion segmentations
from 59 subjects.
For the distribution of data to train, validation and
test, we allocate 45 subjects to train, 5 subjects to va-
lidation and 9 subjects to test data. We use valida-
tion accuracy to determine the number of epochs with
which to train the networks. In the experiments, af-
ter 60 epochs the validation accuracy did not improve
and even started to drop, for this reason we decided to
stick to this number for the training of our networks,
to prevent over-fitting.
As validation measures we consider four different
measures detailed next.
Dice Similarity Coefficient (DSC) is a statistical
overlapping measure that quantifies the similarity be-
tween two segmentations. This measure is between 0
and 1; 0 meaning no similarity and 1 meaning a per-
fect match between segmentations. We compute it as
a percentage as follows.
DSC = 100
2T P
2T P + FP + FN
,
VISAPP 2018 - International Conference on Computer Vision Theory and Applications
264

where TP is the number of True Positives, FP is the
number of False Positives, and FN is the number of
False Negatives.
True Positive Rate (TPR) is the percentage of the le-
sion voxels with respect to the total GT lesion voxels,
which is also called the sensitivity. This measure is
between 0 and 1, and the higher the better although
it has to be considered together with other measures
for the quality of the segmentation. We express it as a
percentage.
T PR = 100
T P
#LV GT
,
where #LVGT is the number of lesion voxels in the
GT.
False Discovery Rate (FDR) is the percentage of
false positive voxels with respect to the output seg-
mentation performed by the method. The measure is
between 0 and 1, and low values are desired. We ex-
press it as a percentage.
FDR = 100
FP
#LV f ound
,
where #LV found is the number of lesion voxels de-
tected by the method.
Volume Difference (VD) is the percentage of the ab-
solute difference between the GT lesion volume and
the volume of the lesions found by the automatic met-
hod with respect to the GT lesion volume. This me-
asure does not give information about the overlap of
the two segmentations but gives an idea about the re-
lative volumes. The minimum and the ideal value for
this measure is 0 but there is no maximum for this me-
asure. 0 value means the lesion volumes in the met-
hod segmentation and the GT are the same in size,
although it might mot mean a perfect overlap. We ex-
press it as a percentage.
V D = 100
k#LV f ound − #LV GT k
#LV GT
.
Moreover, we also consider the computation of
#CC lesion GT: The number of connected compo-
nents (CCs) in the GT segmentation; #CC lesion
found: The number of connected components found;
and #CC lesion coincided: The number of connected
components in the GT that has some overlap with
the connected components in the method segmen-
tation. Note this might be bigger that the number
of connected components found since one CC found
can coincide with multiple CCs in the ground truth.
A Connected Component (CC) can be defined as a
group of lesion voxels connected with each other and
it can be of different sizes.
3.2 Results
Tables 1 and 2 contain the results of the seven appro-
aches considered in the paper together with LST ap-
proach. As can be seen the final model, which is the
3 class model with cascade have the best performance
in all the measures except the TPR. The decrease in
the TPR is understandable, since as we removed the
FP we also had to sacrifice some TP; although in com-
parison it is very small in number. The best model
also surpassed the LST method in all the measures in-
cluding the TPR.
Note that values in Table 2 have to be compared
with # Lesion Voxels in GT (#LV GT) which is 11573
± 9873 and # Lesion CC in GT (#LCC GT) which is
125.6 ± 61.4. As can be seen, the detected number of
lesions is closest to the real number with the 3-class
cascade model, which means the false negatives have
substantially been eliminated. Also with this model,
around 102 CCs are detected, and there is some over-
lap with 72 of the LCCs in the GT, which is the hig-
hest proportion among the models. Although there
are other models with higher LCC coincided with the
GT, these models have an exorbitantly high number of
LCCs found, the majority of which are false detecti-
ons.
Besides analyzing the quantitative results and to
better understand the behavior of the segmentation al-
gorithm on the MRI images, we qualitatively inspect
the results in Figure 3 and Figure 4.
Figure 3 shows the results of the best model in ex-
ample images from 4 subjects of the test set. The ma-
nual segmentation or TP are shown in dark blue, the
FP in light blues, and the FN in pink. As can be seen
from the figure, there is detection around the GT seg-
mentation for most of the lesions and there is a high
amount of overlap but there are also differences. The
differences can be partly due to the subjective nature
of manual MS segmentation or difficulty in obtaining
the real borders, even by a human expert.
Figure 4, which is an up-close version of a re-
gion from Figure 3. It shows the T1 image (left), the
T2 image (middle) and segmentation result (right) of
the same region. As can be seen from the figure, the
manual segmentation is more conservative while our
model is over-segmenting a region as lesion if there
is a corresponding hypo-intensity in T1 and hyper-
intensity in T2. Also note that the GT segmentation is
more jagged and dispersed while the model automa-
tic segmentation is rounder and more connected. This
is expected since the probability of two neighboring
voxels being segmented as lesions both is high since
they have a very similar neighborhood. Based on our
observations, there were also some cases in the GT
Multiple Sclerosis Lesion Segmentation using Improved Convolutional Neural Networks
265
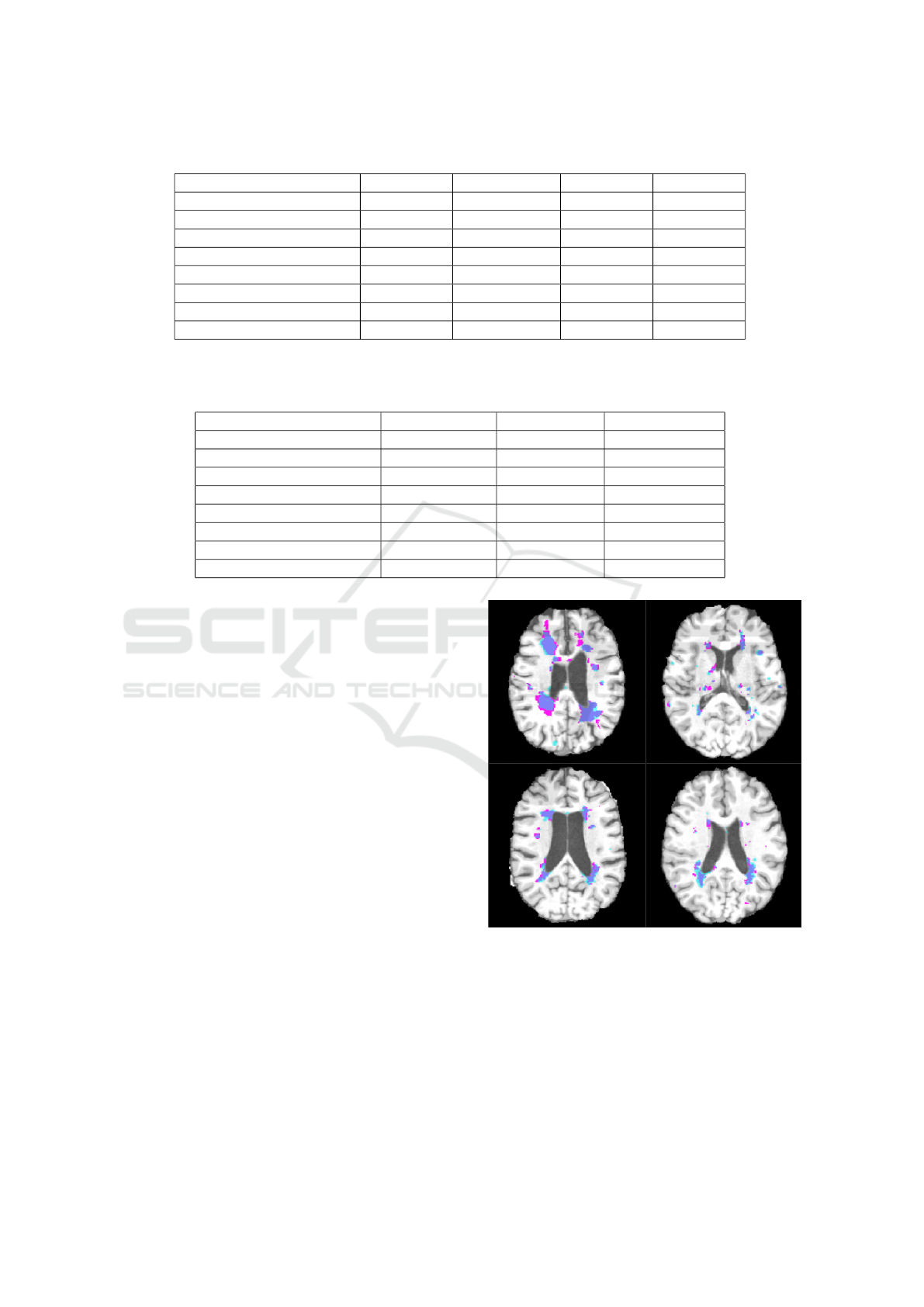
Table 1: Final comparison between models based on DSC, VD, TPR and FDR.
METHOD DSC% VD% TPR% FDR%
LST (LST, 2017) 42.0 ± 19.6 76.7 ± 98.0 49.0 ± 18.3 59.6 ± 22.1
Patch11- w/o location 32.8 ± 21.1 746.5 ± 891.2 81.8 ± 6.6 77.2 ± 16.9
Patch 11- with location 32.7 ± 18.7 650.3 ± 721.1 86.0 ± 9.1 78.2 ± 14.0
Patch 19- with location 45.2 ± 20.1 160.5 ± 212.3 62.5 ± 16.8 60.8 ± 20.3
Patch 11- Cascade 45.3 ± 16.0 178.6 ± 145.7 76.4 ± 14.1 66.2 ± 14.5
Patch 11- 4 class 37.3 ± 16.9 333.9 ± 285.5 79.8 ± 10.3 74.3 ± 13.7
Patch 11 - 3 class 54.8 ± 13.9 39.6 ± 33.8 61.4 ± 14.5 48.2 ± 16.2
Patch 11 - 3 class, Cascade 57.5 ± 12.4 22.8 ± 16.5 59.7 ± 14.6 42.5 ± 14.3
Table 2: Final comparison between models based on # Lesion Voxels found (#LV found), # Lesion CC found (#LCC found)
and # Lesion CC which coincide in GT and found (#LCC coincide). Note that # Lesion Voxels in GT is 11573 ± 9873 and #
Lesion CC in GT is 125.6 ± 61.4.
METHOD #LV found #LCC found #LCC coincided
LST (LST, 2017) 12936 ± 10231 25.3 ± 9.4 31.8 ± 16.6
Patch11- w/o location 36848 ± 11170 561.3 ± 128.6 92.3 ± 41.6
Patch 11- with location 38765 ± 18541 474.9 ± 90.3 91.9 ± 42.2
Patch 19- with location 17468 ± 13073 151.7 ± 62.6 51.6 ± 29.5
Patch 11- Cascade 24695 ± 19621 113.8 ± 38.8 69.9 ± 37.0
Patch 11- 4 class 30826 ± 20973 257.1 ± 42.7 85.1 ± 37.6
Patch 11 - 3 class 13190 ± 11721 150.1 ± 51.8 75.2 ± 38.0
Patch 11 - 3 class, Cascade 11981 ± 10986 101.7 ± 50.4 72.7 ± 31.5
that was contrary to the MS lesion definition, which
caused some ”wrong” FN in our results and this might
be explained with some special case, a human error or
an error in the alignment process of the images. More
detailed qualitative results can be seen from Figure 6.
From our observations, the model seems to cap-
ture the hypo-intensity in T1 and hyper-intensity in
T2 technically but misses some of the intuitions, dom-
ain knowledge or the subjectivity of the human ex-
pert. The lesions are detected by the proposed mo-
del in hypo-intense regions in T1 and hyper-intense
regions in T2 and the borders are determined by T1
hypo-intense region, which is the correct behavior.
Another positive property of the automatic seg-
mentation is that in the majority of the lesions there
is some detection on the lesion or very close to the
lesion, although the overlapping is far from perfect.
For instance, if seen from a cross-section, there is a
GT lesion that seems not detected, when we advan-
ced a few voxels up or down along the perpendicular
axis of the cross-section, we observe, in most cases, a
lesion detected by the model. This could be due to the
difficulty in determining the real borders of a lesion.
In Figure 5 we can see that the number of auto-
matically detected lesion voxels increased with the
number of GT lesion voxels. This means that the seg-
mentation result of our model is indicative of the real
lesion load. This information can be used to see the
progression of the lesions in an MS patient.
Figure 3: Segmentation result, with the best model, of four
subjects in axial plane. TP: Dark Blue, FP: Light Blue, FN:
Pink. Better visualization in pdf.
In terms of computational cost, we observe that
changing the patch size from 11x11x11 to 19x19x19
increases the computational time to more than twice
its original value (approximately, 6 hours as oppo-
sed to 14 hours in the AWS configuration (Amazon,
2017) we chose), but proportionally less than the in-
crease in the number of initial features, which is:
VISAPP 2018 - International Conference on Computer Vision Theory and Applications
266

Figure 4: From left to right: T1, T2 and Segmentation re-
sult, close-up from a lesion region. Better visualize in pdf.
Figure 5: The number of Ground Truth lesions vs. the num-
ber of lesions found.
(19/11)
3
≈ 5.2. The application of the cascade, on
the other hand, increases the computational time ap-
proximately to twice its original value, which is ex-
pected since we train the same size CNN with the
same number of samples twice (additionally the time
for evaluating the training set with the first model,
which is negligible in comparison). Increasing the
class size, however, has less dramatic effect on the
computational time causing comparatively a smaller
increase than twofold.
4 CONCLUSIONS AND FUTURE
WORK
In this paper, we have studied several CNN models for
the MS lesion segmentation problem. We have started
with model definitions from similar approaches in the
literature and we have developed our final proposal
by adding new design decisions: defining a new sub-
sampling process, including lesion location informa-
tion in the CNN input, considering a 3-classes classi-
fication problem and adding a two-stage cascade. We
have shown that these improvements have a positive
effect on the final performance. We have shown that
using DL with an appropriate design, we can define a
method that learns the general rule for MS lesion seg-
mentation. We have obtained promising results for
all the validation measures on a real dataset of 59 pa-
tients from Hospital Cl
´
ınic. Moreover, the results re-
present significant improvement over the commonly
used automatic method of LST. The proposed mo-
del have been able to detect the lesion regions, which
have different intensity values in MRI with respect to
their neighborhood, meaning that it has captured the
mathematical relationship between a 3D patch and the
class of its center voxel to a certain degree. Although
it has captured the general relationship, it has failed to
learn some exceptions requiring domain knowledge
that are applied by human experts during MS lesion
segmentation. This limitation could be due to the fact
that there is not enough cases in the training set for
such exceptions or that it is necessary to feed more
information to the CNN (e.g. more context) for the
model to capture these patterns.
As future work we plan to add more information
about the context and brain spatial dependencies in
order to improve the results. Although we have ad-
ded the location information to add some context, this
may not have been enough since the training set may
not be representative of the exceptions. Thus, adding
the resulting lesion/non-lesion label information (de-
tected by the model) of the neighboring voxels in the
evaluation of a voxel may help achieve better classifi-
cation. In order to include these spatial dependencies,
we can consider Conditional Random Fields as a se-
cond stage.
In the literature, Restricted Boltzmann Machines
(RBM) or auto-encoders have been used to obtain an
initial representation of the data, which could lead to
better classifiers. This type of unsupervised methods
can also take advantage of unlabeled data. Therefore,
it might be a good idea to start with an RBM or auto-
encoder and apply our models subsequently.
Finally, another improvement may come from in-
creasing the number of MRI modalities during trai-
ning. We used T1, T2 but adding more modalities
such as FLAIR, fMRI, diffusion MRI could give more
information about the nature of a voxel.
ACKNOWLEDGEMENTS
This work was partly supported by the Ministry of
Economy, Industry and Competitiveness of Spain
(TIN2015-66951-C2-1-R and TIN2013-47008-R), by
Catalan Government (2014-SGR-1219 and SGR-
1187), the EU H2020 and a NVIDIA GPU Grant.
Multiple Sclerosis Lesion Segmentation using Improved Convolutional Neural Networks
267
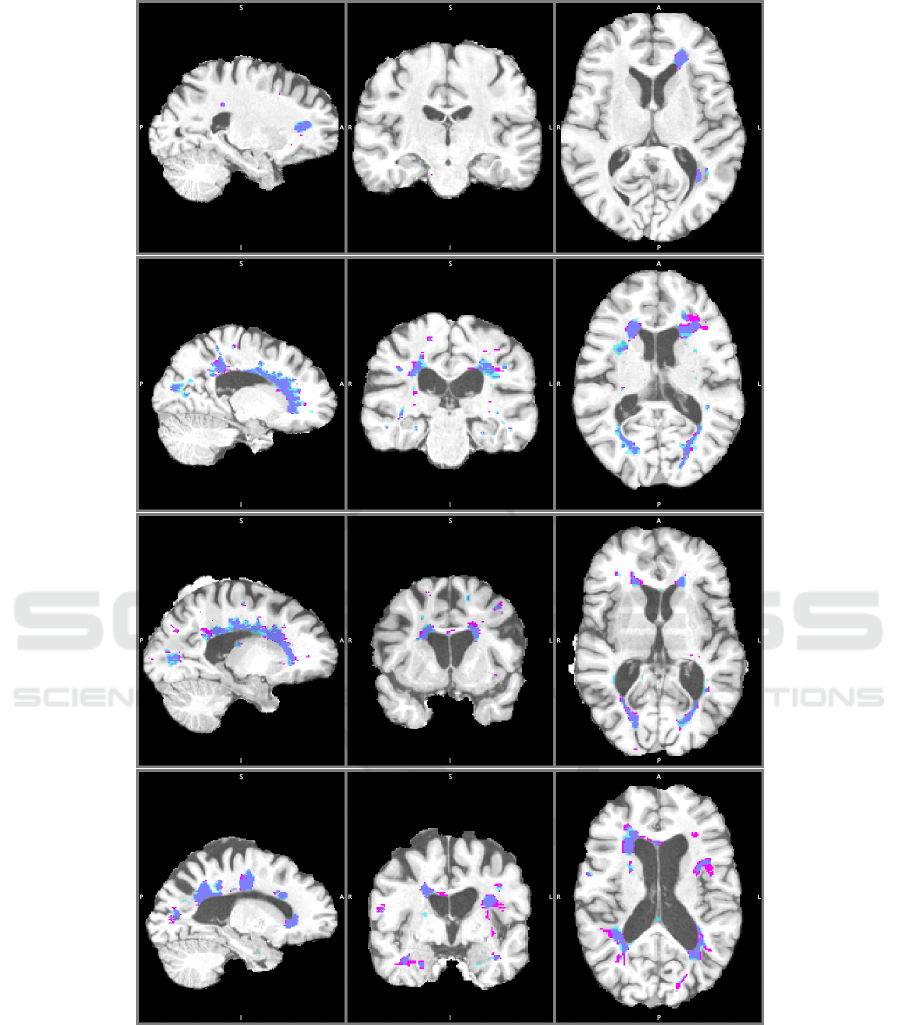
Figure 6: Other segmentation results, with the best model, of four subjects in saggital, coronal and axial plane. TP: Dark
Blue, FP: Light Blue, FN: Pink. Better visualization in pdf.
REFERENCES
Havaei, M., Guizard, N., Larochelle, H. and Jodoin, P-M.
(2016). Deep learning trends for focal brain pathology
segmentation in MRI. arXiv:1607.05258
Greenspan, H., Van Ginneken, B. and Summers,R. M.
(2016). Deep Learning in Medical Imaging: Over-
view and Future Promise of an Exciting New Techni-
que. IEEE Transactions on Medical Imaging, 35 (5),
pp. 1153-1159, 2016.
Garcia-Lorenzo, D., Francis, S., Narayanan, S., Arnold, D.
L., Collins, D. L. (2013). Review of automatic seg-
VISAPP 2018 - International Conference on Computer Vision Theory and Applications
268

mentation methods of multiple sclerosis white matter
lesions on conventional magnetic resonance imaging.
Medical Image Analysis, 17 (1), pp. 1-18, 2013.
Vaidya, S., Chunduru, A., Muthu Ganapathy, R., Krish-
namurthi, G. (201). Longitudinal Multiple Sclerosis
Lesion Segmentation Using 3D Convolutional Neural
Networks. IEEE International Symposium on Biome-
dical Imaging, New York, April 2015.
Brosch, T., Yoo, Y., Tang, L.Y., Li, D.K., Traboulsee, A.,
Tam, R. (2015). Deep convolutional encoder networks
for multiple sclerosis lesion segmentation. Medical
Image Computing and Computer-Assisted Interven-
tion (MICCAI) 2015, pp. 3-11. Springer.
Brosch, T., Tang, L., Yoo, Y., Li, D., Traboulsee, A. (2016).
Deep 3d convolutional encoder networks with short-
cuts for multiscale feature integration applied to mul-
tiple sclerosis lesion segmentation. IEEE Transactions
on Medical Imaging, 35 (5), pp. 1229-1239, 2016.
Ghafoorian, M., Karssemeijer, N., et al. (2017). Location
sensitive deep convolutional neural networks for seg-
mentation of white matter hyperintensities. Scientific
Reports 7 (1): 5110, 2017.
Valverde, S., Cabezas, M., et al. (2017). Improving auto-
mated multiple sclerosis lesion segmentation with a
cascaded 3D convolutional neural network approach.
NeuroImage, 155, pp. 159-168, 2017.
Plis, S. M., Hjelm, D. R., Salakhutdinov, R., Allen, E. A.,
Bockholt, H. J., Long, J. D., Johnson, H. J., Paulsen, J.
S., Turner, J. A. and Calhoun, V. D. (2014). Deep le-
arning for neuroimaging: a validation study. Frontiers
in Neuroscience, 8, 2014
Goodfellow, I., Bengio, Y., Courville, A. (2016). Deep le-
arning. MIT Press Book.
Bengio, Y. (2012). Practical recommendations for gradient-
based training of deep architectures Neural Networks:
Tricks of the Trade, pp. 437-478, 2012.
National MS Society. http://www.nationalmssociety.org/
Deep Learning. http://deeplearning4j.org/
Freesurfer for processing and analyzing brain MRI images.
https://surfer.nmr.mgh.harvard.edu/
MS Lesion Segmentation Tool (LST). http://
www.statistical-modelling.de/lst.html
Neuroimaging in Python (Nibabel). http://nipy.org/nibabel/
Scikit-learn pre-processing package. http://scikit-learn.org/
stable/modules/preprocessing.html
Amazon Web Services. https://aws.amazon.com/
Multiple Sclerosis Lesion Segmentation using Improved Convolutional Neural Networks
269
