
ECG-based Biometrics using a Deep Autoencoder for Feature Learning
An Empirical Study on Transferability
Afonso Eduardo, Helena Aidos and Ana Fred
Instituto de Telecomunicac¸
˜
oes, Instituto Superior T
´
ecnico, Universidade de Lisboa, Lisbon, Portugal
afonso.eduardo@tecnico.ulisboa.pt, {haidos, afred}@lx.it.pt
Keywords:
Biometrics, User Identification, Electrocardiogram (ECG), Deep Learning, Feature Learning, Transfer
Learning, Deep Autoencoder.
Abstract:
Biometric identification is the task of recognizing an individual using biological or behavioral traits and, re-
cently, electrocardiogram has emerged as a prominent trait. In addition, deep learning is a fast-paced research
field where several models, training schemes and applications are being actively investigated. In this paper, an
ECG-based biometric system using a deep autoencoder to learn a lower dimensional representation of heart-
beat templates is proposed. A superior identification performance is achieved, validating the expressiveness
of such representation. A transfer learning setting is also explored and results show practically no loss of
performance, suggesting that these deep learning methods can be deployed in systems with offline training.
1 INTRODUCTION
1.1 Biometric Identification Systems
Biometric identification systems have been a topic of
great research interest in the past 50 years (Jain et al.,
2016). These systems use biological and behavioral
traits, as opposed to more traditional identification
methods, such as those based on tokens or knowledge
(e.g. passwords).
A typical biometric identification system is, as
depicted in Figure 1, comprised of two stages: en-
rollment and identification. In the former, features
extracted from the pre-processed acquired signal,
known as templates, are stored in a database along
with the corresponding identification labels. In the
latter, the biometric signal is processed in a similar
fashion, apart from the labels that are unknown, and,
as such, the system performs recognition by using the
information available in the stored templates.
Recent advances in this area focus on adaptative
systems, motivated by the interpretation of many bio-
Figure 1: A general biometric identification system.
metric trait features as being generated from non-
stationary stochastic processes. Naturally, the pres-
ence of the latter leads to performance degradation
as the templates acquired during the enrollment stage
can become poor representatives of the biometric
traits to be recognized. Primers on this subject can be
found in (Roli et al., 2008) and (Rattani et al., 2015).
1.2 Biometric Traits and ECG
A significant body of work has been devoted to an-
alyze the suitability of different traits for biometric
recognition according to a number of criteria which
include uniqueness, universality, performance and ac-
ceptability (Jain et al., 2016).
Typical traits are fingerprint, face and iris. How-
ever, motivated by (Biel et al., 2001), electrocardio-
gram (ECG) as a biometric trait has also gained trac-
tion within the research community. Indeed, in this
seminal work, it is empirically shown that ECG (see
Figure 2) can contain sufficient information as to al-
low person recognition. The ECG-based biometrics
literature has been growing ever since: different ac-
quisition experiments have been reported as well as
different feature extraction and classification models.
In-depth literature surveys can be found in (Odinaka
et al., 2012) and, more recently, in (Fratini et al.,
2015).
Eduardo, A., Aidos, H. and Fred, A.
ECG-based Biometrics using a Deep Autoencoder for Feature Learning - An Empirical Study on Transferability.
DOI: 10.5220/0006195404630470
In Proceedings of the 6th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2017), pages 463-470
ISBN: 978-989-758-222-6
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
463

Figure 2: A normal ECG (single heartbeat) with annotated
P-QRS-T waves.
1.3 Deep Learning
1.3.1 Overview
One field that has gained a tremendous amount of at-
tention among machine learning researchers and prac-
titioners alike is that of deep learning, or deep neural
networks (LeCun et al., 2015). In essence, these mod-
els are comprised of simple nonlinear transformation
modules that are arranged in a network whose topol-
ogy is often problem-specific and can be interpreted
as the prior belief of the modeller on how the data
should interact. These building blocks can, in turn, be
stacked or used with other modules to build a network
of many layers.
Despite the infancy of research exploring deep
learning theoretical properties (for recent efforts, see
(Eldan and Shamir, 2016) and (Cohen et al., 2016)),
the intuition is that these models can learn more ab-
stract and thus expressive representations of the in-
put data at each layer (Goodfellow et al., 2016).
This type of learning, also known as representa-
tion/feature/manifold learning, is indeed a useful
property for deep learning methods to have, because
traditional machine learning systems require exten-
sive domain knowledge and careful feature engineer-
ing in order to find a suitable representation that can
be fed to the next learning module, usually a classifier.
And, although initially deployed in computer vision
applications, deep learning techniques have also been
applied to other domains such as natural language
processing and time series, namely physiological data
(L
¨
angkvist et al., 2014). For instance, in (Martinez
et al., 2013), a Convolutional Neural Network (CNN)
automates the feature extraction process in an affect
detection model.
Another research topic that is gaining momentum
within the deep learning community is that of trans-
fer learning. It has been established that many mod-
els require a significant amount of training data, in
addition to a training process that can itself be ex-
pensive. These causes justify the need to study how
transferable the features learned by deep networks
might be (Yosinski et al., 2014). For instance, a po-
tential scenario of transfer learning would be one in
which the model is trained on a base dataset, different
from the possibly smaller or constantly evolving tar-
get dataset. If the features to be learned are suitable
to both datasets, it is asserted to work favorably, en-
abling the use of deeper networks without the risk of
overfitting and bypassing the often costly full training
procedure.
1.3.2 Autoencoders and Related Work
Autoencoder (AE), another type of neural network,
has also found some applications. Summarily, this
model is a self-supervised technique that is trained
to attempt to copy its input to its output (Goodfellow
et al., 2016). AE has been used to learn lower dimen-
sional representations of the original data and to pre-
train other deep learning networks, e.g. CNNs. The
greedy layer-wise pretraining process has, however,
gradually been replaced in favor of better initializa-
tion weights, e.g. (Glorot and Bengio, 2010), or dif-
ferent training schemes, e.g. (Srivastava et al., 2015).
Nevertheless, AE and its variants are still found in re-
cent publications whose focus is on ECG.
In (Xiong et al., 2015), a Denoising AE (DAE)
is used for ECG signal enhancement and, in (Rahhal
et al., 2016), it is used to pretrain a Deep Neural Net-
work (DNN) for active classification of ECG signals.
In (Del Testa and Rossi, 2015), ECGs are compressed
via DAE. This lossy compression architecture is then
compared against other popular algorithms according
to compression ratio, reconstruction fidelity and com-
putational complexity. The authors conclude that AE
can achieve a high compression efficiency and a small
reconstruction error. It is also stated that the energy
consumption is low and thus suitable for deployment
in wearable devices.
ECG-based recognition using neural networks is
not a novel idea, e.g. (Shen et al., 2002) and (Wan and
Yao, 2008). The use of deep learning tools in a range
of applications is, however, relatively recent. That
said, to the best of these authors’ knowledge, the only
publication whose focus is simultaneously on ECG-
based recognition and deep learning is that of (Page
et al., 2015). In this work, the authors explore a DNN
architecture in an authentication setting using a public
database. The latter consisting of roughly 300 1-lead
ECG recording sessions obtained from 90 volunteers
in a resting state. The authors report an Equal Error
Rate (EER) of 0.0582%.
ICPRAM 2017 - 6th International Conference on Pattern Recognition Applications and Methods
464

1.4 Contributions
In this paper, an ECG-based biometric identification
system where a deep autoencoder is used for feature
learning is proposed and tested in a similar setting
than that of (Carreiras et al., 2016), i.e. a single chan-
nel ECG bioemtric system is evaluated on data ob-
tained from a local hospital and whose subjects are
not necessarily in a resting state.
Additionally, it is shown the deep autoencoder
successfully learns a projection into a lower dimen-
sional space and that using such data representa-
tion leads to a superior identification performance.
This also highlights the versatility of deep learn-
ing methods as compressed and possibly privacy-
preserving representations arise naturally; qualities
that are paramount in a a number of applications, in-
cluding those of mobile healthcare.
Another consideration is that of transfer learning.
In ECG-based biometrics, it is not unusual to have a
small target dataset. For instance, if users were to be
enrolled in the system based on a 10-second frame,
assuming a heart rate of 60 beats per minute, at most
10 good-quality heartbeat traces could be acquired. In
a small-scale deployment, this would lead to a small
training dataset which in turn would put constraints
on the type of deep learning models that could be
learned as to avoid overfitting. In addition, if the tar-
get dataset were to be constantly evolving due to the
unenrollment of old and enrollment of new users, the
training of the deep networks would need to be re-
peated in a naive implementation. One can however
imagine how costly or even prohibitive it would be
in practical environments, especially so in decentral-
ized embedded applications. For these reasons, it is
assessed the performance degradation that the system
would incur if the feature extraction module (deep au-
toencoder) were to be trained on a different dataset,
i.e. on traces belonging to subjects that would not be
enrolled in the system.
2 METHODOLOGY
2.1 System Overview
The proposed model can be summarily described as
a one-to-many template matching system whose tem-
plates are given by an encoded representation of the
individual ECG heartbeats. The mapping function
is, in turn, learned by the encoder submodule of the
deep autoencoder and its hyperparameters, such as its
topology, are selected by using a validation set.
Furthermore, to determine the effectiveness of this
representation, the proposed model is compared with
a similar system where the templates are not encoded,
the latter of which is henceforth referred as baseline
or, in short, as B.
From a practical standpoint, it is also assumed that
a dataset with the following characteristics is avail-
able: D
0
=
{
(x
i
,y
i
)
}
N
i=1
, where x
i
∈ R
d
is a (heart-
beat) template, with d 1; and, y
i
∈ C
0
⊂ N is the
corresponding label, i.e. subject identifier, with C
0
de-
noting the set of possible labels. Details on how this
dataset might be built can be found in Section 3.1; its
analysis is, however, beyond the scope of this paper.
Similarly, the classifier is characterized in Section 3.2.
2.2 Deep Autoencoder
Before proceeding with the brief description of au-
toencoders, the reader is assumed to already be fa-
miliar with the basic concepts of neural networks and
backpropagation. Additional details and clarifications
can be found in (Goodfellow et al., 2016). It should
also be mentioned that the authors find difficult to
identify the point at which an autoencoder is to be
considered a deep autoencoder. That said, in this
work, an autoencoder is said to be deep, if the number
of hidden layers is greater than one.
An autoencoder is a special type of feedforward
neural network whose purpose is to learn how to re-
construct the inputs belonging to a given dataset X
?
:
X = [{x : x ∈ X
?
⊂ X }]
T
. It is generically comprised
of two submodules: the encoder function, λ : X 7→ Z,
with Z = [{z : z = λ(x),∀x ∈ X
?
}]
T
being the encoded
inputs; and the decoder function, ψ : Z 7→ X . In addi-
tion, an objective function (the reconstruction loss),
L(X,λ,ψ), must be defined in order to update the
function parameters, i.e. the weights and biases of
the autoencoder network, via backpropagation. For
instance, for real-valued inputs a squared error is
typically employed: L(X , λ,ψ) = ||X −
ˆ
X||
2
F
, where
ˆ
X = [{ ˆx : ˆx = (ψ ◦ λ)(x), ∀x ∈ X
?
}]
T
denotes the re-
constructed inputs.
Moreover, in order for the autoencoder to learn a
useful representation of the input data and avoid the
identity function, it might be necessary to perform
regularization by adding constraints. This can be done
explicitly by designing a network with a bottleneck,
i.e a network whose hidden layers have less units than
those at visible layers. In this case, the autoencoder
is said to be undercomplete (see Figure 3) because
it learns an undercomplete representation of the data.
Alternatively, a more subtle approach can be adopted:
regularization terms can be added to the loss function
to promote sparsity; or other techniques, such as data
ECG-based Biometrics using a Deep Autoencoder for Feature Learning - An Empirical Study on Transferability
465

Figure 3: Schematic of an undercomplete deep autoencoder.
corruption (Vincent et al., 2008) or dropout (Srivas-
tava et al., 2014), can be used. In this case, the au-
toencoder is also able to learn an overcomplete repre-
sentation provided that the space of the encoded data
Z is allowed to be higher dimensional than that of the
original, X . In this work, only the former approach,
i.e. a network with bottleneck, is explored and the
hyperparameter space is described in Section 3.2.
2.3 Learning Schemes
In order to evaluate the proposed biometric system in
the transfer learning setting mentioned in Section 1,
there is the need to create the base and target datasets,
both similar to one another.
The next subsection describes the procedure that
generates subsets from D
0
, followed by the specifica-
tion of the different deep autoencoder schemes whose
performances, in addition to B, are to be compared in
Section 3.3.
2.3.1 Data Generating Process
Let φ
1
: R
d
× N 7→ R
d
× N denote a function such
that D
1
= φ
1
(D
0
) with D
1
⊂ D
0
. First, φ
1
draws
|C
0
|/2 samples without replacement from C
0
, gener-
ating C
1
and, subsequently, for each c ∈ C
1
, n dif-
ferent random templates and their corresponding la-
bels are selected, ensuring that the resulting dataset
D
1
has no class bias. Correspondingly, there ex-
ists a function φ
2
: R
d
× N 7→ R
d
× N such that
D
2
= φ
2
(D
0
), D
2
⊂ D
0
, and whose C
2
= C
0
\C
1
. Note
that D
1
∪ D
2
⊂ D
0
and D
1
∩ D
2
=
/
0. Each of these
datasets is then split into train, validation and test,
D
i
=
D
train
i
,D
val
i
,D
test
i
, i ∈ {1,2}, by randomly
leaving one sample per class for validation and an-
other for test.
Furthermore, let D
3
= D
1
∪D
2
with D
test
3
= D
test
1
.
The remainder subsets, training and validation, follow
D
train
3
∪ D
val
3
= D
3
\ D
test
3
, with D
val
3
being generated
by randomly selecting one sample per class.
2.3.2 Schemes
Three scenarios are investigated and are as follows:
M1. The feature extraction module, i.e. deep au-
toencoder, is trained and validated on D
train
1
and D
val
1
respectively. The learned function,
λ
1
: R
d
7→ R
p
with p < d, is then applied to
all, N
1
, templates in D
1
, resulting in the en-
coded dataset. The latter of which is given
by Λ
1
=
{
(z
1i
,y
i
)
}
N
1
i=1
=
Λ
train
1
,Λ
val
1
,Λ
test
1
,
with z
1i
= λ
1
(x
i
),∀x
i
∈ D
1
. The classifier is
trained on Λ
train
1
∪ Λ
val
1
and tested on Λ
test
1
.
M2. The deep autoencoder learns λ
2
: R
d
7→ R
p
by training and validating on D
train
2
and D
val
2
respectively. Similarly, λ
2
is applied to each
template in D
1
, giving rise to Λ
2
. The clas-
sifier is trained on Λ
train
2
∪ Λ
val
2
and tested on
Λ
test
2
.
M3. The deep autoencoder learns λ
3
: R
d
7→ R
p
by training and validating on D
train
3
and D
val
3
respectively. As a result, Λ
3
is obtained by
applying λ
3
to each template in D
1
. The
classifier is trained on Λ
train
3
∪Λ
val
3
and tested
on Λ
test
3
.
Notice that D
1
is considered to be the target
dataset. Therefore, M1 corresponds to the typical sce-
nario in which the feature extraction module is trained
on instances from the same dataset, whereas a differ-
ent dataset is used in M2. Further, note that the com-
parison that follows in Section 3.3 is fair because both
datasets are structurally equivalent, i.e. with respect
to the number of training and validation instances and
number of classes. On the other hand, M3 addresses
the case where instances from both datasets are used,
adding insight on how the system might behave if
additional data are available. Finally, for complete-
ness, it should be mentioned that, in B, the classifier
is trained on D
train
1
∪ D
val
1
and tested on D
test
1
.
3 EXPERIMENTS
3.1 Dataset
The data, as in (Carreiras et al., 2016), were collected
from a local hospital using a Philips PageWriter Trim
III device, a sampling rate of 500 Hz with 16 bit
resolution and a 12-lead placement. A total of 960
10-second records, amounting to 709 different sub-
jects, was hand labeled by an expert and classified
as being normal, i.e. with no apparent pathologies.
To ensure coherence, records acquired on days other
ICPRAM 2017 - 6th International Conference on Pattern Recognition Applications and Methods
466
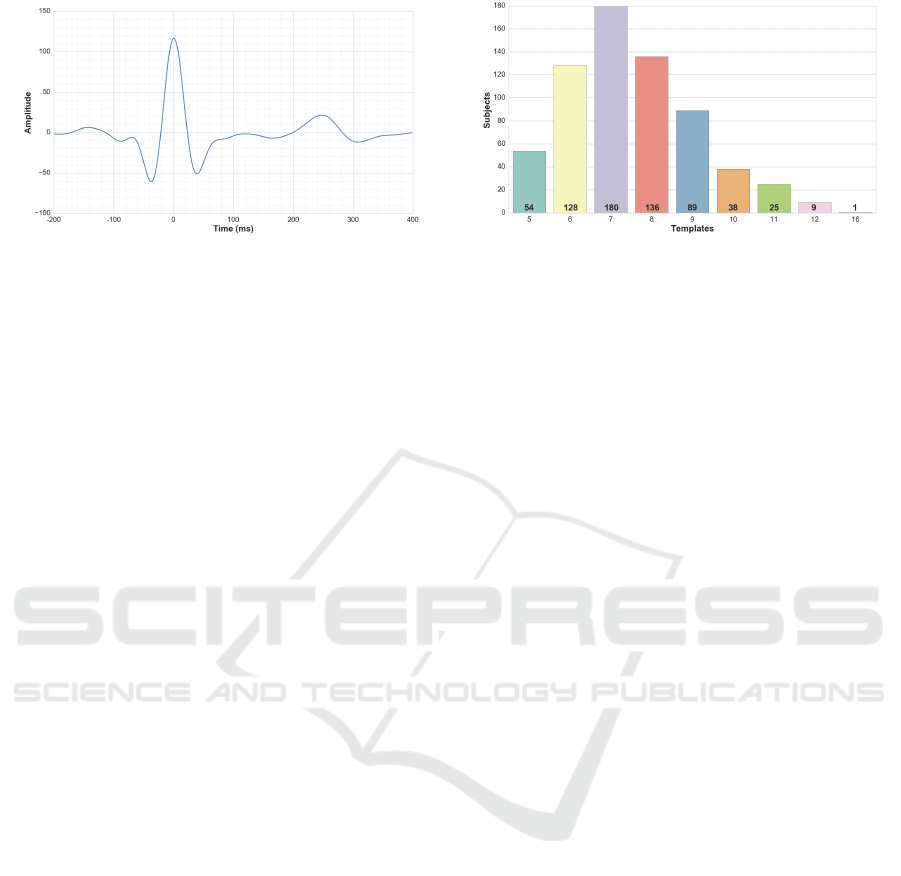
(a) An example of a heartbeat template. (b) Histogram.
Figure 4: Description of the dataset D
0
.
than the first were discarded because these sessions
were not taken at regular intervals and the number of
subjects with more than one session was considered
insufficient for this study to be carried.
In order to remove the baseline wander and other
possible noise sources, the same procedure reported
in (Marques et al., 2015) was followed. The raw sig-
nals were filtered by a 150th-order bandpass Finite
Impulse Response (FIR) filter with lower and higher
cutoff frequencies of 5 and 20 Hz, respectively. Sub-
sequently, the resulting signals were transformed into
heartbeat templates by taking a fixed-length window
[−200,400] ms around the detected R peaks.
Abnormal templates were then removed using
DMEAN, a method proposed by (Lourenc¸o et al.,
2013), with parameters α = 0.5, β = 1.5 and Eu-
clidean distance. Similarly, subjects whose mean dis-
tances to the respective mean wave template were
over the upper fence, i.e. Q3 + 1.5 × (Q3 − Q1),
where Q1 and Q3 are the first and third quartiles, were
classified as outliers and discarded, giving rise to the
dataset D
0
.
The dataset can be described as D
0
=
{
(x
i
,y
i
)
}
N
i=1
,
where x
i
∈ R
d
with d = 300; y
i
∈ C
0
≡ {1,...,U} with
U = 660; and N = 4966. A x
i
template and the his-
togram of this dataset are shown in Figure 4.
3.2 Parameters
In this section, the parameters of the experiments are
described. These correspond to the parameter n in the
data generating process (see Section 2.3.1), the classi-
fier and the hyperparameters of the deep autoencoder.
As shown in Figure 4(b), the minimum number of
templates per subject in D
0
is 5. Therefore, to take
full advantage of D
0
, i.e. to take all subjects into con-
sideration, in the data generating process, n = 5.
Regarding the classifier, the same algorithm is
used across all schemes, including B. The model is a
k-Nearest Neighbor classifier with k = 3, cosine dis-
tance and a voting scheme that follows the majority
rule with the (first, according to a given ordering)
nearest neighbor being the tiebreaker.
In the autoencoder, a range of different hyperpa-
rameters are used and the system selects, each time, a
potentially different set of hyperparameters based on
identification performance, i.e. the design that leads
to the least number of misclassified instances in the
validation set. Four different topologies are consid-
ered and are the following: [300, 100, 50, 100, 300],
[300, 150, 50, 150, 300], [300, 150, 75, 50, 75, 150,
300], [300, 150, 100, 50, 100, 150, 300]. Note that all
of these can be interpreted as performing a lossy com-
pression whose ratio is 6:1. The possible activation
functions are tanh and ReLU, except in the last layer,
whose function is only allowed to be tanh. In addition,
tied weights are not considered and their initialization
is the same as reported in (Glorot and Bengio, 2010).
Each deep autoencoder is optimized via Adam with
default α,β
1
,β
2
,ε, (Kingma and Ba, 2015), and shuf-
fled minibatches of size 256. The objective function
is the mean squared error without any regularization
terms. Different training epochs are considered, rang-
ing from 100 to 500 with steps of size 100. Finally,
before being fed to the autoencoder, all templates are
scaled to [-1, 1]. This step is min-max normalization
with the minimum and the maximum being given by
the minimum and maximum values in the training set
and to whose values a 10% buffer is added.
The implementation is written in Python and,
in addition to the SciPy stack (Jones et al., 2001),
Theano (Theano Development Team, 2016) and
Keras (Chollet, 2015) are used.
3.3 Results and Discussion
To evaluate the performance of B, M1, M2 and M3, a
total of 1000 trials are conducted. In each trial, an in-
stance from the data generating process is drawn and
the resulting identification errors (one per scheme),
i.e. the ratios of failed over total number of attempts,
on the instantiated test set (330 subjects) are recorded.
ECG-based Biometrics using a Deep Autoencoder for Feature Learning - An Empirical Study on Transferability
467

(a) Identification error: boxplots with annotated medians.
1
3
4
2
M1
(2.13)
M3
(1.71)
M2
(2.16)
B
(4.00)
Critical Difference: 0.180
(Minimal difference to achieve statistical significance)
(b) Scheme comparison via Nemenyi test.
Figure 5: Identification performance.
These experiments are conducted under the simplify-
ing assumption of a closed world, i.e. all subjects are
assumed to be enrolled in the system and, therefore,
any identification attempt gives rise to a match. Note,
however, that this does not compromise the main ob-
jective which is to evaluate the expressiveness of the
representation learned by the deep autoencoder un-
der different learning schemes. A boxplot summary
is shown in Figure 5(a).
In addition, statistical tests, proposed by (Dem
ˇ
sar,
2006), are performed to determine whether the dif-
ferences among B, M1, M2 and M3 are significant.
A Friedman rank sum test rejects the hypothesis that
all methods have equivalent performance at α = 0.01
with p < 10
−20
. Pairwise comparisons are then car-
ried with a Nemenyi test at a 99% confidence level.
Results are summarized in Figure 5(b) with the aver-
age rank across datasets of each scheme being shown.
At this confidence level, schemes whose pairwise dis-
tances are greater than the critical distance are consid-
ered to have a performance difference that is signifi-
cant. This is true for all pairs, except for (M1, M2).
Three major observations can be drawn from these
results. First, the representations learned by the deep
autoencoder under any scheme lead to a superior iden-
tification performance when compared to B. This dif-
ference is significant and suggests that ECG heartbeat
data lie in a lower dimensional nonlinear embedding,
a space where the classification task is made easier.
For instance, intuitively, the information contained in
the QRS complex alone is of great importance and its
original representation is in a highly redundant form.
Notice also that the representation is learned in an un-
supervised fashion in that the autoencoder, by defi-
nition, is not given the task of learning a discrimi-
nant space. Learning such space might, however, fur-
ther increase the identification performance and can
be achieved by adding a classification layer and fine-
tuning the network parameters, giving rise to a stan-
dard DNN.
Another observation is that the performance error
distributions of M1 and M2 are almost identical, a fact
that is underlined by the not statistically significant
average rank difference. Thus, it is proved that the
type of transfer learning under discussion is appro-
priate, i.e. it is possible to use a base dataset, differ-
ent from the target, to train the deep learning model,
provided that the ECG signals are acquired and pre-
processed in a similar fashion. This, in turn, opens
several possibilities as these models can be trained
offline and deployed in a plethora of environments,
ranging from small to large-scale deployments, in-
cluding embedded applications.
Finally, M3 achieves the best performance among
the four schemes. Such significant increase cannot,
however, be attributed to the usage of samples from
the target dataset during training and validation, since
the previous observation dismisses this proposition.
The increase is due to the availability of larger train-
ing and validation sets. In particular, whereas the
training/validation sets in both M1 and M2 only have
990/330 samples, 2310/660 samples are available in
M3. Naturally, performance can be improved by gath-
ering additional samples, but this task might prove to
be difficult, especially so for biomedical data. Data
augmentation appears thus as a possible avenue. Data
ICPRAM 2017 - 6th International Conference on Pattern Recognition Applications and Methods
468

can be artificially generated by applying techniques
that range from simple translations or scaling to more
elaborate procedures. For instance, a state-space gen-
erative model capturing the ECG dynamics can be
employed (McSharry et al., 2003), (Sameni et al.,
2007). Alternatively, more general generative mod-
els can be learned, such as those based on variational
autoencoders (Kingma and Ba, 2014) or on generative
adversarial networks (Goodfellow et al., 2014).
4 CONCLUSION AND FUTURE
WORK
This paper proposed an ECG-based biometric system
where lower dimensional nonlinear representations of
heartbeat templates are learned via deep autoencoder.
The expressiveness of these representations were as-
sessed and results show that they lead to a superior
identification performance. Additionally, a transfer
learning setting was evaluated, i.e. a scheme where a
base dataset, different from the target, is used to train
the autoencoder. This learning scheme is shown to
have a similar performance to that of a scheme where
the same target dataset is used during training. A
result that opens several possibilities for deep learn-
ing methods in ECG-based biometrics, including their
deployment in wearable devices and other embedded
applications.
On a side note, the authors would like to mention
the importance of publicly available data in research,
namely in ECG-based biometrics. The scarcity of
large public ECG databases specifically designed for
identification purposes and whose data are collected
over multiple sessions is hindering the advancement
of this research field. The authors are aware of the
privacy and legal framework governing medical data.
Nevertheless, joint endeavors should be made to ad-
dress this problem.
For future work, and in addition to the sugges-
tions stated in Section 3.3, a more exhaustive search in
the hyperparameter space can be performed. This in-
cludes regularization techniques and different topolo-
gies. Interesting practical studies would be on identi-
fication performance with varying feature dimension-
ality, number of subjects and templates. Different pre-
processing methods can also be investigated such as
those based on denoising autoencoders or state-space
signal processing using Bayesian filtering. Similarly,
other types of classifiers can be employed. Finally,
different deep learning approaches can be explored,
using, for instance, spectrograms and CNNs.
ACKNOWLEDGEMENTS
This work was supported by the Portuguese Founda-
tion for Science and Technology, scholarship num-
ber SFRH/BPD/103127/2014 and grant PTDC/EEI-
SII/7092/2014. In addition, the authors would like to
thank Diana Batista (IT/IST, Portugal) for her work
on ECG segmentation.
REFERENCES
Biel, L., Pettersson, O., Philipson, L., and Wide, P. (2001).
ECG analysis: a new approach in human identifica-
tion. IEEE Transactions on Instrumentation and Mea-
surement, 50(3):808–812.
Carreiras, C., Lourenc¸o, A., Silva, H., Fred, A., and Fer-
reira, R. (2016). Evaluating Template Uniqueness in
ECG Biometrics. In Filipe, J., Gusikhin, O., Madani,
K., and Sasiadek, J., editors, Informatics in Control,
Automation and Robotics: 11th International Con-
ference, ICINCO 2014 Vienna, Austria, September
2–4, 2014 Revised Selected Papers, pages 111–123.
Springer International Publishing.
Chollet, F. (2015). Keras. https://github.com/fchollet/keras.
Cohen, N., Sharir, O., and Shashua, A. (2016). On the Ex-
pressive Power of Deep Learning: A Tensor Analysis.
JMLR Workshop and Conference Proceedings, 49:1–
31.
Del Testa, D. and Rossi, M. (2015). Lightweight Lossy
Compression of Biometric Patterns via Denoising
Autoencoders. IEEE Signal Processing Letters,
22(12):2304–2308.
Dem
ˇ
sar, J. (2006). Statistical Comparisons of Classifiers
over Multiple Data Sets. Journal of Machine Learning
Research, 7:1–30.
Eldan, R. and Shamir, O. (2016). The Power of Depth for
Feedforward Neural Networks. JMLR Workshop and
Conference Proceedings, 49:1–34.
Fratini, A., Sansone, M., Bifulco, P., and Cesarelli,
M. (2015). Individual identification via electrocar-
diogram analysis. Biomedical engineering online,
14(1):78.
Glorot, X. and Bengio, Y. (2010). Understanding the
difficulty of training deep feedforward neural net-
works. Proceedings of the 13th International Con-
ference on Artificial Intelligence and Statistics (AIS-
TATS), 9:249–256.
Goodfellow, I., Bengio, Y., and Courville, A. (2016). Deep
learning. Book in preparation for MIT Press.
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B.,
Warde-Farley, D., Ozair, S., Courville, A., and Ben-
gio, Y. (2014). Generative adversarial nets. In Ad-
vances in Neural Information Processing Systems 27
(Proceedings of NIPS).
Jain, A. K., Nandakumar, K., and Ross, A. (2016). 50 Years
of Biometric Research: Accomplishments, Chal-
lenges, and Opportunities. Pattern Recognition Let-
ters, 79:1–28.
ECG-based Biometrics using a Deep Autoencoder for Feature Learning - An Empirical Study on Transferability
469

Jones, E., Oliphant, T., Peterson, P., et al. (2001).
SciPy: Open source scientific tools for Python.
https://www.scipy.org/.
Kingma, D. and Ba, J. (2014). Auto-Encoding Variational
Bayes. In 2nd International Conference on Learning
Representations (ICLR2014).
Kingma, D. and Ba, J. (2015). Adam: A method for
stochastic optimization. In 3rd International Confer-
ence on Learning Representations (ICLR2015).
L
¨
angkvist, M., Karlsson, L., and Loutfi, A. (2014). A re-
view of unsupervised feature learning and deep learn-
ing for time-series modeling. Pattern Recognition Let-
ters, 42(1):11–24.
LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep learn-
ing. Nature, 521(7553):436–444.
Lourenc¸o, A., Silva, H., Carreiras, C., and Fred, A. (2013).
Outlier detection in non-intrusive ecg biometric sys-
tem. In AIMI International Conf. on Image Analysis
and Recognition - ICIAR, pages 43–52.
Marques, F., Carreiras, C., Lourenc¸o, A., Fred, A., and Fer-
reira, R. (2015). Ecg biometrics using a dissimilarity
space representation. In Proceedings of the Interna-
tional Conference on Bio-inspired Systems and Signal
Processing (BIOSTEC 2015), pages 350–359.
Martinez, H. P., Bengio, Y., and Yannakakis, G. (2013).
Learning Deep Physiological Models of Affect. IEEE
Computational Intelligence Magazine, Special Issue
on Computational Intelligence and Affective Comput-
ing, 8(2):20–33.
McSharry, P. E., Clifford, G. D., Tarassenko, L., and Smith,
L. A. (2003). A dynamical model for generating syn-
thetic electrocardiogram signals. IEEE Transactions
on Biomedical Engineering, 50(3):289–294.
Odinaka, I., Lai, P.-H., Kaplan, A. D., O’Sullivan, J. A.,
Sirevaag, E. J., and Rohrbaugh, J. W. (2012). ECG
Biometric Recognition: A Comparative Analysis.
IEEE Transactions on Information Forensics and Se-
curity, 7(6):1812–1824.
Page, A., Kulkarni, A., and Mohsenin, T. (2015). Utilizing
deep neural nets for an embedded ECG-based biomet-
ric authentication system. IEEE Biomedical Circuits
and Systems Conference: Engineering for Healthy
Minds and Able Bodies, BioCAS 2015 - Proceedings,
pages 0–3.
Rahhal, M. M. A., Bazi, Y., Alhichri, H., Alajlan, N., Mel-
gani, F., and Yager, R. R. (2016). Deep learning ap-
proach for active classification of electrocardiogram
signals. Information Sciences, 345:340–354.
Rattani, A., Roli, F., and Granger, E., editors (2015). Adap-
tive Biometric Systems. Advances in Computer Vision
and Pattern Recognition. Springer International Pub-
lishing.
Roli, F., Didaci, L., and Marcialis, G. L. (2008). Adaptive
Biometric Systems That Can Improve with Use. In
Ratha, N. K. and Govindaraju, V., editors, Advances in
Biometrics: Sensors, Algorithms and Systems, pages
447–471. Springer London.
Sameni, R., Shamsollahi, M. B., Jutten, C., and Clifford,
G. D. (2007). A nonlinear Bayesian filtering frame-
work for ECG denoising. IEEE Transactions on
Biomedical Engineering, 54(12):2172–2185.
Shen, T. W., Tompkins, W. J., and Hu, Y. H. (2002). One-
lead ecg for identity verification. In Engineering in
Medicine and Biology, 2002. 24th Annual Conference
and the Annual Fall Meeting of the Biomedical Engi-
neering Society EMBS/BMES Conference, 2002. Pro-
ceedings of the Second Joint, volume 1, pages 62–63
vol.1.
Srivastava, N., Hinton, G. E., Krizhevsky, A., Sutskever,
I., and Salakhutdinov, R. (2014). Dropout : A Sim-
ple Way to Prevent Neural Networks from Overfit-
ting. Journal of Machine Learning Research (JMLR),
15:1929–1958.
Srivastava, R. K., Greff, K., and Schmidhuber, J. (2015).
Training very deep networks. In Advances in Neu-
ral Information Processing Systems 28 (Proceedings
of NIPS), volume 28, pages 2377–2385.
Theano Development Team (2016). Theano: A Python
framework for fast computation of mathematical ex-
pressions. arXiv e-prints, abs/1605.02688.
Vincent, P., Larochelle, H., Bengio, Y., and Manzagol, P.-
A. (2008). Extracting and composing robust features
with denoising autoencoders. In Proceedings of the
25th International Conference on Machine Learning
(ICML-08).
Wan, Y. and Yao, J. (2008). A Neural Network to Identify
Human Subjects with Electrocardiogram Signals. In
Proceedings of the world congress on engineering and
computer science.
Xiong, P., Wang, H., Liu, M., and Liu, X. (2015). De-
noising Autoencoder for Eletrocardiogram Signal En-
hancement. Journal of Medical Imaging and Health
Informatics, 5(8):1804–1810.
Yosinski, J., Clune, J., Bengio, Y., and Lipson, H. (2014).
How transferable are features in deep neural net-
works? Advances in Neural Information Processing
Systems 27 (Proceedings of NIPS), 27:1–9.
ICPRAM 2017 - 6th International Conference on Pattern Recognition Applications and Methods
470
