
Laboratory Information Management System for NGS Genomics Labs
Jitao Yang
1,2
, Shuai Zhu
2
, Guangpeng Xu
2
and Chao Han
2
1
School of Information Science, Beijing Language and Culture University, Bejing 100083, China
2
Annoroad Gene Technology, Beijing 100176, China
Keywords:
LIMS, NGS, Genome Sequencing, Genomics, Sample Tracking.
Abstract:
The goal of genome sequencing is to unravel the ordered sequence of nucleic acids that form the DNA or RNA
of a given sample. Genome sequencing lab requires the ability to select and track a large amount of samples
through many experimental steps. Therefore, laboratory information management system (LIMS) is needed
to provide a way of automating the laboratory experimental procedures and track the samples. LIMSs have
been proposed and developed for many years, but still remain difficult for labs to implement successfully. In
this paper, we demonstrate our genomic next generation sequencing (NGS) LIMS solution. We developed a
web-based LIMS with flexible configuration and customization for NGS laboratories, and can help laboratories
track samples and optimize experimental procedures and business workflows. We also describe our solution of
integrating LIMS with the existing enterprise business information systems. Finally, we share our experience
for the implementation of a successful LIMS.
1 INTRODUCTION
Genome next generation sequencing is now com-
monly adopted, and has a broad areas of applica-
tions, such as the Non-invasive Prenatal DNA Testing,
ctDNA Testing for Non-invasive Tumor Personalized
Therapy, Plant and Animal Molecular Breeding, Ge-
netics and Evolution, Microorganism and Ecological
Environment, etc.
To manage tens of thousands of samples that are
subject to NGS analysis, it is inevitable to develop ad-
equate laboratory information management system to
track and manage the NGS workflows. However, it is
extremely difficult to manage the NGS expriment and
analysis procedures accurately and efficiently, given
the issues of multiple-source samples enrolling, na-
tional and international logistical management and
tracking, fragmented procedures for assessment and
processing of samples, the intricacies of molecular
experimental steps, and the complex and multiple
pipelines of NGS processing.
LIMS provides many benefits for the users of lab-
oratory, several of the main benefits identified are out-
lined below:
• brings accuracy and accessibility to the flow of
samples and data in laboratory,
• universally accessible data via the web rather than
digging through files,
• years of data can be kept and queried conve-
niently,
• business efficiency improvement,
• data quality control,
• efficient sample tracking and management,
• automated and in-depth customer reports,
• integration with laboratory instruments,
• experimental steps quality control,
• building automated analysis pipelines,
• status and results sharing with collaborators and
clinicians,
• financial management,
• access control,
• track and analyze trends,
• error reduction.
In this paper, we demonstrate our genomic NGS
LIMS solution. Our LIMS is a web-based system
with flexible configurations and customizations for
NGS laboratories and can support the integration of
multiple NGS instruments, it can help laboratories
track samples and optimize experimental procedures
and business workflows. We also give solution of in-
tegrating LIMS with the existing enterprise business
information systems. Finally, we describe the most
326
Yang J., Zhu S., Xu G. and Han C.
Laboratory Information Management System for NGS Genomics Labs.
DOI: 10.5220/0006149503260333
In Proceedings of the 10th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2017), pages 326-333
ISBN: 978-989-758-213-4
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

likely causes for a failed LIMS and share our experi-
ence for the implementation of a successful LIMS.
The rest of the paper is structured as follows: Sec-
tion 3 introduces the design of LIMS, while Section 4
describes the implementation of LIMS, and Section 5
presents the integration of LIMS with existing infor-
mation systems. Section 6 discusses the implemen-
tation of a successful LIMS. Section 7 concludes the
paper and outlines future work.
2 RELATED WORKS
Some commercial and open source LIMSs (Bath
et al., 2011; PerkinElmer, 2016; Grimes and Ji, 2014;
Progeny, 2016; Illumina, 2016) are available but typ-
ically require extensive modification and extension to
address the specific needs of NGS genomics labs. We
therefore developed a web-based LIMS, which is ro-
bust and flexible for managing the samples and the
NGS processes.
3 SYSTEM DESIGN
As described in Figure 1, the functionalities of our
LIMS can be grouped into the following categories:
• Enrollment of Sample Information. Enrollment
of sample information is not a simple form fill-
ing process but constituted by two or even three
steps, and supports multiple business models. For
instance, salesman who stationed at hospitals will
input the basic sample information (e.g., sam-
ple code, sample type, photo of sample sheet)
into system by mobile applications embedded in
Wechat. Then, when the sample along with the
sample sheet are transported to company, typists
in company will finish the full sample information
(in sample sheet) enrollment. Finally, the correct-
ness of sample information enrolled in LIMS will
be checked by another team in company.
• Sample Logistics Information Tracking. Samples
come from different cities distributed in the coun-
try, and will be transported to our head quarter to
be tested. Each sample logistics information will
be tracked by system. When samples are packed
and sent to our company, package logistics code
will be scanned into system. Then, the system
can get the newest logistics information from ex-
press companies through their open APIs. We
can check the package logistics information status
during the whole transportation process. If some-
thing unusual (e.g., delayed, destroyed) happened
in logistics process, the recipients and senders can
response quickly to take steps to minimize the
losses.
• Sample Assessment and Processing. Depends on
sample attribute, sample sheet, sample number,
test type or more factors, the samples arrived at
company can be divided to different processing
directions including rejection, resampling, stor-
age, or flow to the experiment steps. Sample re-
cipients, experiment operators and genomic ana-
lysts can choose the processing directions and op-
erate in the system.
• Experimental Management. Experimental man-
agement is the most important and complicated
part of LIMS. Based on sample type, test type,
previous processing result and operator’s subjec-
tive judgment, the samples will experience multi-
ple experimental steps such as plasma isolation,
nucleic acid extraction, molecular library con-
struction, molecular library quality control, and
etc. Our system is also flexible to support the con-
figuration of different experimental processes.
• Sequencing of Samples. After the experimental
steps, the samples are ready to be sequenced by
NGS instrument (e.g., Illumina HiSeq X Ten). To
minimize the cost of using sequencing reagents,
generally, one NGS instrument operation will be
required to sequencing as many samples as pos-
sible, therefore, samples belonging to different
test business line will be combined together in the
pooling process. After sequencing, the raw .bcl
data will be separated into deferent genomic anal-
ysis business processes. Abnormal result data will
be auto labeled by particular rules configured.
• Genomic Analysis. Before genomic analysis,
LIMS will prepare the raw .bcl data for the fil-
ter and quality control software to be processed.
The generated FASTQ data will be further pre-
pared for the genomic analysis pipelines operated
in the cluster computing environment. After ge-
nomic analysis, LIMS will retrieve the analysis
result set to system so as to wait for the genetic
interpretation.
• Genetic Interpretation. Sequencing and analysis
results are presented in a neat and orderly man-
ner in LIMS for the genetic interpretation scien-
tists. Additionally, LIMS can provide relevant
knowledge base for genetic interpretation such
as gene-disease associations from several public
data sources (e.g. AutDB (Mindspec, 2016), Dis-
GeNET (Pinero et al., 2015), OMIM (Hamosh
et al., 2015)) and the literatures.
Laboratory Information Management System for NGS Genomics Labs
327

Enrollment of
Sample
Information
Sample
Logistics
Tracking
Sample
Assessment and
Processing
Experimental
Management
Sequencing of
Samples
Genomic
Analysis
Genetic
Interpretation
Master Data
Management
Instrument
Management
Project
Management
Query and
Statistics
System Management
Report
Management
Quality Control
Figure 1: The Application Architecture of LIMS.
• Report Management. The genetic interpretation
result will be classified by the report management
module and transferred into different report gen-
eration pipelines of the reporting system.
• Quality Control Management. Quality control
exists in the whole NGS work flows, including
experiment steps quality control, reagents qual-
ity control, sequencing data quality control, ge-
nomic analysis quality control, genetic interpre-
tation quality control, and etc. Different quality
control processes can also be configured in sys-
tem.
• Master Data Management. Master data manage-
ment module includes tens of thousands of fun-
damental data for the operation of LIMS, such as
the details data for experiment templates, barcode
rules, agent information and so on. This module
is usually maintained by department manager or
system administrator, because master data include
very important and sensitive data of the company.
• Instrument Management. Instrument manage-
ment module not only records instrument prop-
erty information like barcode, type, number, loca-
tion, status, application methods and so on, but
also associates instruments to a certain experi-
ment step. An experiment step including its as-
sociated instruments could be exported as a detail
guide which is very helpful for the operators.
• Project Management. Project management mod-
ule provides the functions for project managers
a convenient way to manage and monitor the
projects, including project defining, project status
monitoring, process intervening, and so on.
• Query and Statistics. Query and statistics mod-
ule provides powerful functions of advanced data
query and visualized multidimensional data statis-
tic and analysis. Instead of querying through mil-
lions of data items to find meaningful results in
experimental workflows, researchers can quickly
identify the information of their interests through
this module.
• System Management. System management mod-
ule provides the features of user management, role
management, privilege management, department
management, business management, organization
management. A user’s role and his privileges
could configured flexibly.
4 SYSTEM IMPLEMENTATION
Our LIMS is written in Java using the Spring (Piv-
otal, 2016), SpringMVC, Mybatis (Goodin et al.,
2016) web application framework and implementa-
tion is platform-independent. Our web servers run
Linux/CentOS and we use the MySQL relational
database management system. The system architec-
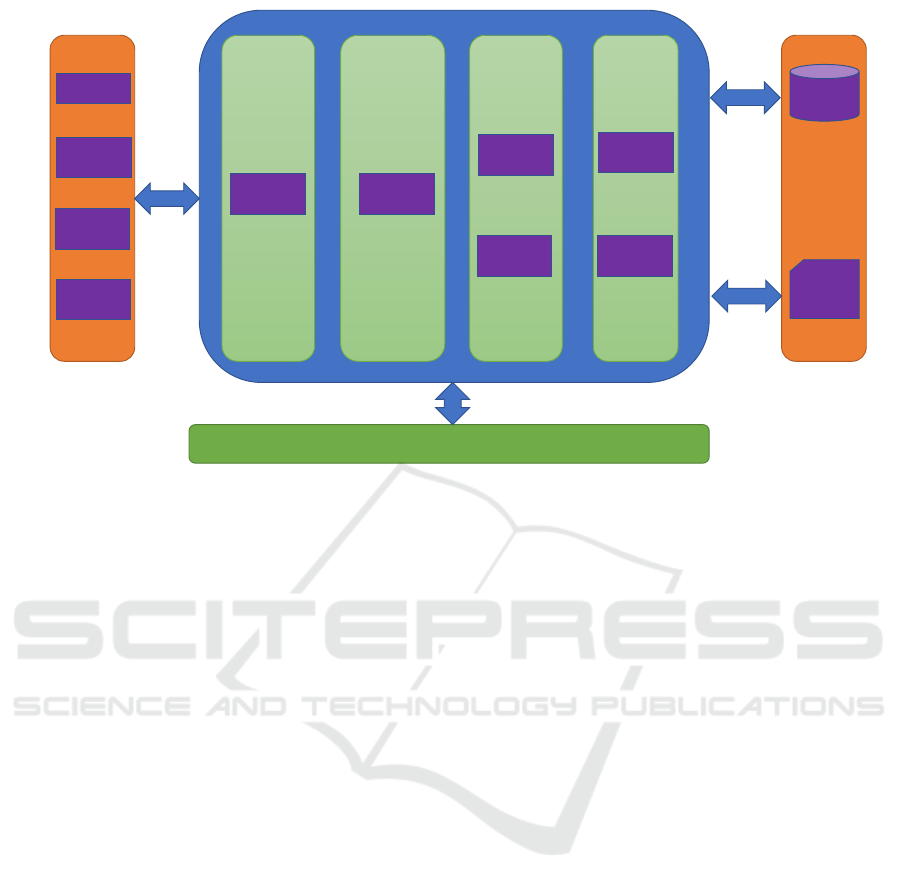
ture of LIMS is described in Figure 2.
The web interface is designed to handle a variety
of functions in a modular format. The left column
includes the function category of business process.
The top navigation bar provides advanced manage-
ment and analysis functions.
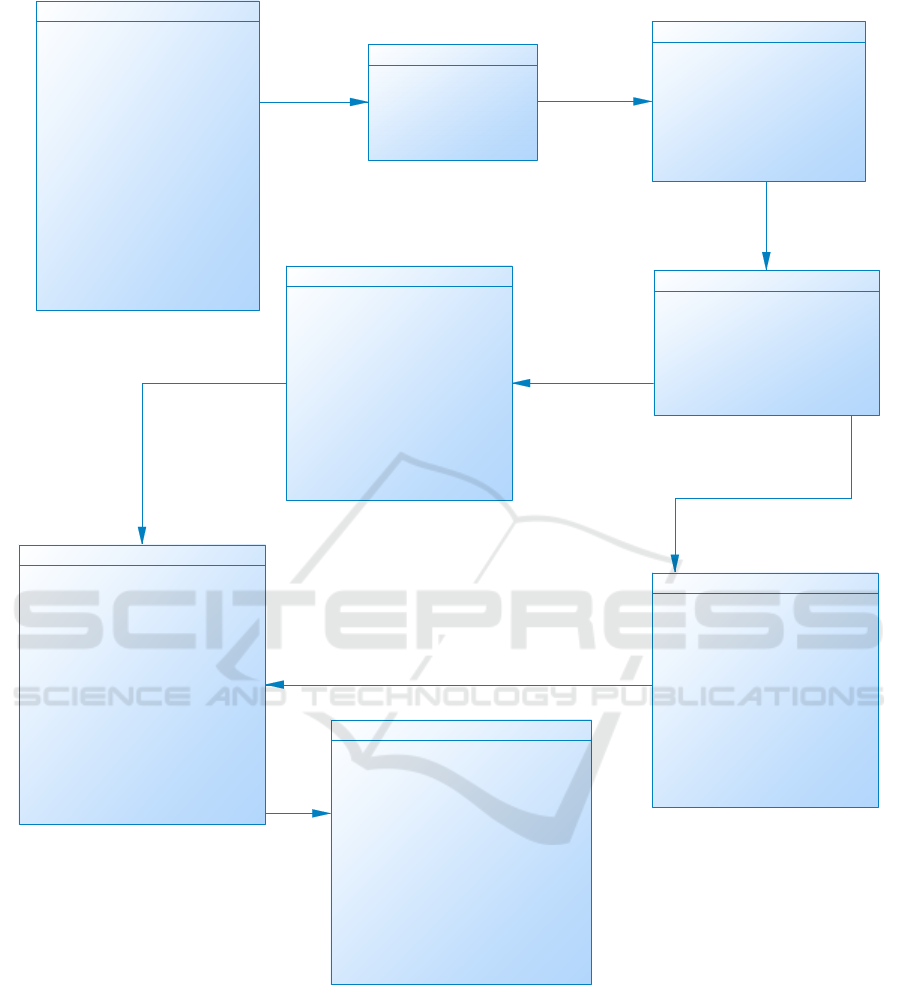
We have more than one hundred data tables in the
system, Figure 3 shows part of our database schema
for data tables.
HTTPS (Hypertext Transfer Protocol Secure) is
supported and is implemented for the system. User’s
access to functionality is controlled via user roles
HEALTHINF 2017 - 10th International Conference on Health Informatics
328
Data
Access
(DAO)
Business
Process
(Service)
Data
Control
(Controller)
View
CRM and other enterprise information systems
Oracle,
MySQL
File
System
MyBatis/
JDBC
I/O
IOS
Android
WAP
PC
HTTP
JSP
Spring
MVC
Business
Service
Web
Service
DAO
File
Process
Web Service
Figure 2: The System Architecture of LIMS.
which are defined and managed by the system man-
agement module.
Our LIMS currently supports the following Il-
lumina NGS sequencing platforms: HiSeq X Ten,
MiSeq, HiSeq, HiSeq 4000, HiSeq2500, NextSeq
500/550AR, and could be configured for other type
of NGS instruments.
5 INTEGRATION WITH
EXISTING SYSTEMS
Implementation of a LIMS requires a good degree of
integration with the existing business information sys-
tems in enterprise. We integrate LIMS to our two ex-
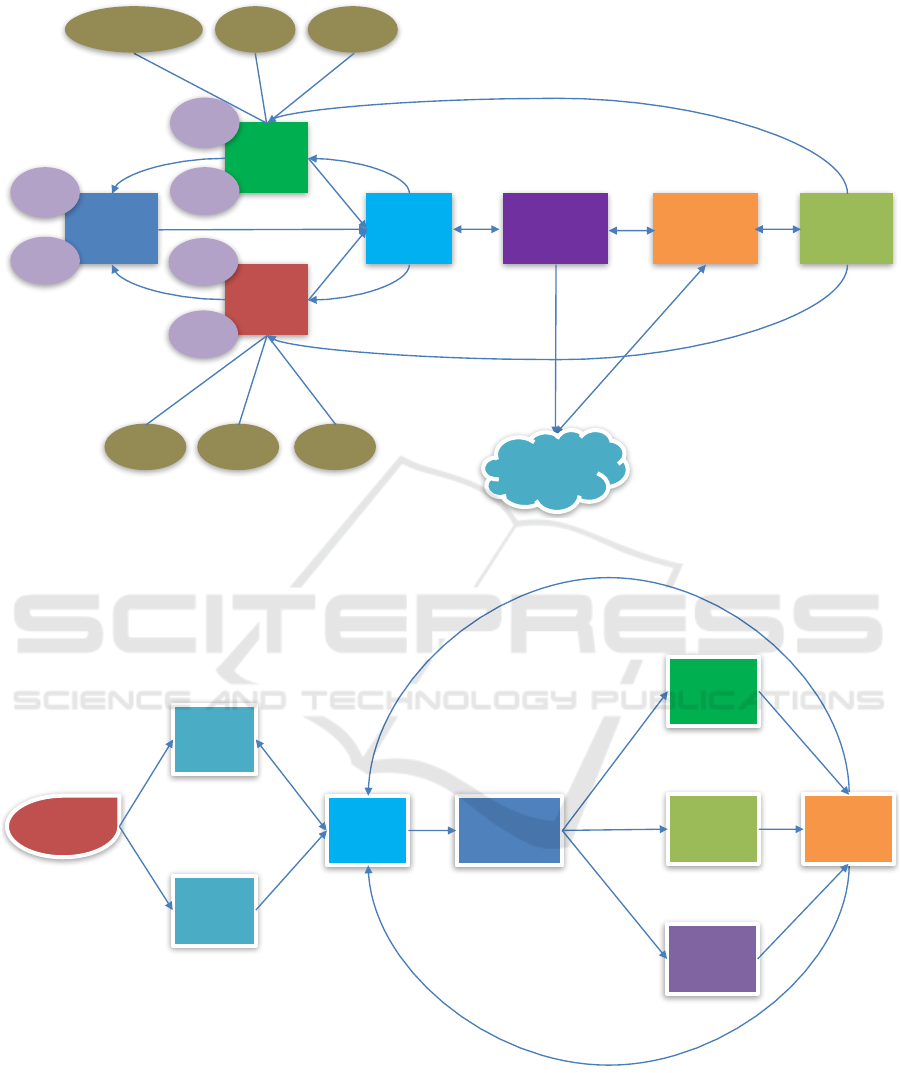
isting business system platforms as demonstrated in
Figure 4 and Figure 5.
5.1 Integrating with Company Systems
Figure 4 describes the business process in our com-
pany.
Customer Relationship Management (CRM) sys-
tem serves primarily our clinical and health ser-
vices, including Personal Genome Test, NIPT (Non-
Invasive Prenatal DNA Testing), Cancer Gene Ther-
apy, and etc. CRM supports sample information
collection, hospital information management, agent
management, product management, sales manage-
ment, sample management, doctor management, fi-
nancial management, customer reports management,
and etc.
Project Management (PM) system serves primar-
ily our technical services, including the research area
of genomics, transcriptomics, epigenetics, and etc.
PM includes the functions of sample information
enrollment, contract management, project schedule
management, project establishment, job order man-
agement, quality control, research achievement man-
agement, and etc.
Logistics system provides the management func-
tions of express package, express company, alert time,
receipt time, samples, logistics tracking, and etc.
All the information related to NGS experiments
and analysis will be synchronized into LIMS, there-
fore, the sample information could be enrolled
through:
• CRM web portal, or
• CRM mobile app, or
• PM web portal, or
• PM mobile app, or
• LIMS directly.
The sequencing result data will be stored in Net-
work Attached Storage (NAS) system, and then fil-
tered and classified to be analyzed by hundreds of dif-
ferent genomic analysis pipelines.
The genomic analysis result data will then flow to
the reporting system, which can produce hundreds of
different areas of professional reports automatically.
The reporting system will finally distribute the
user reports to CRM and PM, then the user can receive
Laboratory Information Management System for NGS Genomics Labs
329
1 / 1
SampleInfo
sampleCode
testType
sampleType
hospital
outpatientCode
bloodTime
patientName
age
weight
gestationalWeeks
phoneNumber
mailingAddress
lastMenstruation
bUltrasound
trisome
inspectionDoctor
diagnosis
inspectionSheet
medicalHistory
varchar
varchar
varchar
varchar
varchar
date
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
UnboxCheck
unboxCode
description
courierNumber
recipient
dateReceived
status
varchar
varchar
varchar
varchar
date
varchar
PlasmaSeparation
taskLIstCode
recipient
dateReceived
bloodCode
experimentalNumber
plasmaCode
nextFlow
volume
result
varchar
varchar
date
varchar
varchar
varchar
varchar
varchar
varchar
LibraryConstruction
taskListCode
commander
orderTime
plasmaNumber
experimentalNumber
libraryNumber
originalSampleNumber
user
varchar
varchar
date
varchar
varchar
varchar
varchar
varchar
2100Test
taskCode
commander
orderTime
flow
libraryNumber
executionNumber
storageLocation
user
batchNumber
fragmentLength
quality
ratio
originalSampleNumber
molality
varchar
varchar
date
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
QPCRTest
taskCode
commander
orderTime
flow
libraryNumber
executionNumber
storageLocation
user
batchNumber
fragmentLength
massConcentration
ratio
originalSampleNumber
molality
varchar
varchar
date
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
Polling
taskCode
commander
orderTime
sequencingType
sequencingPlatform
sequencingReadLength
serialNumber
libraryNumber
executionNumber
storageLocation
originalSampleNumber
mixingRatio
conversionFactor
convertedConcentration
dilutionRatio
dilutedConcentration
mixingVolume
varchar
varchar
date
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
NGS
taskCode
commander
orderTime
FCNumber
reagentName
instrumentNumber
sequencingType
poolingNumber
libraryVolume
HybVolume
dilutedConcentration
degeneratedConcentration
denaturedSampleVolume
expectedDensity
NGSoperateDate
NGSStopTime
varchar
varchar
date
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
varchar
date
date
Figure 3: Part of Database Schema for LIMS.
their reports through our mobile app or web portal.
We have also constructed a hybrid cloud comput-
ing platform SolarGenomics (SolarGenomics, 2016),
because our business can produce around 10 TB se-
quencing data everyday, both the local computing
cluster and the local distributed storage can not pro-
vide computing and storage resources sufficiently and
elastically.
5.2 Integrating with Hospital Systems
Figure 5 describes the business process in our collab-
orating hospitals.
We collaborate with more than 2000 hospitals. In
some of the hospitals, we set up NGS laboratory and
install our NGS instrument there so that the hospitals
will have the abilities of NGS experiments, sequenc-
HEALTHINF 2017 - 10th International Conference on Health Informatics
330
PM
CRM
LIMS
Genomic
Analysis
Reporting
System
Logistics
NGS
Instruments
app
web
web
1
Cloud
NIPTHealth Cancer
app
web
app
Genomics
EpigeneticsTranscriptomics
Figure 4: The Business Process System Platform in Enterprise.
HIS
LIMS
Cloud
Genomic
Analysis
NGS
Instruments
Samples
data
data
reports
reports
1
Manual
Reporting
System
Head
Quarter
data
Figure 5: The Business Process System Platform in Hospital.
ing, genomic analysis, and genetic interpretation.
The samples in our collaborating hospitals could
be enrolled to LIMS either manually or synchronized
from the hospital information system (HIS). The sam-
ples will be processed by multiple experimental steps
in the laboratory of hospital and then be sequenced by
the NGS instrument placed in the hospital.
Further, the sequencing data will be:
• analyzed by our genomic analysis pipelines in-
stalled in the local hospital computing servers, or
Laboratory Information Management System for NGS Genomics Labs
331

• uploaded to our SolarGenomics cloud computing
platform then analyzed by our genomic analysis
pipelines on cloud, or
• transfered to our head quarter then analyzed by
our bioinformatics scientists.
Finally, our reporting system will give profes-
sional reports to the clinical doctors or patients.
6 A SUCCESSFUL LIMS
IMPLEMENTATION
A large number of LIMS implementations failed to
meet the user’s initial expectations, and this can be
due to the lack of proficient user requirements speci-
fications, the frequent requirements changes, and the
technology-based shortcomings.
The absence of adequate requirements is the
biggest reason why a LIMS may fail, as generally the
software developers are seldom have sufficient knowl-
edge of experiments and NGS, the success of a LIMS
relies on the deep understanding of laboratory and ge-
nomic analysis business needs.
Frequent requirements changes is another reason
causing the fail of LIMS, as the laboratory staffs are
normally focused on a specific area of experiment, it
is hard for them to identify clearly how a LIMS is go-
ing to fit into their laboratory situation and to propose
a general function architecture of LIMS. Addition-
ally, the lab staffs lack the IT knowledge and skills,
they will think the modification of system functions
as a simple task, therefore, the requirements will be
modified frequently. This will postpone the deliver of
project and even fail to publish LIMS.
Technology-based shortcomings should be em-
phasized, as the information technology department
is often not the first class citizen in gene technology
organizations.
Implementation of a LIMS project will also re-
quire a good degree of integration with the exist-
ing enterprise business information systems, data ex-
change mode and standard must be adequately ad-
dressed.
Mapping out clearly the requirements specifica-
tions, controlling the requirements changes effec-
tively, addressing the problem of integrating LIMS
into existing enterprise systems, relying on a strong
IT team will stand a quite high chance of implement-
ing LIMS successfully.
7 CONCLUSIONS
To meet the needs of managing tens of thousands
of samples for genome sequencing, we developed a
web-based laboratory information management sys-
tem that is flexible to be configured to adapt to next
generation sequencing technologies. A LIMS system
is critical to the accurate and effective management
of sample information, experimental data, genome se-
quencing data, and the reproducible analysis results.
Our LIMS addresses all of these needs and seamless
integrates with the existing systems in enterprise and
in collaborating hospitals.
In our LIMS, all the data are stored into distributed
authoritative repositories, samples are traceable from
the enrollment, transportation, experiment, a sequenc-
ing run, quality control, genomic analysis, genetic in-
terpretation, generation of report, to sample storage,
and all the other processing steps in between. In con-
junction with a sample identifier (QR code) encod-
ing rules and advanced query and analysis capabil-
ities, LIMS can quickly identify sample and signif-
icantly reduce errors in the whole steps of business
process. Our LIMS provides a comprehensive and ef-
ficient management solution for NGS genomics labs.
LIMS is very complicated and difficult to imple-
ment, especially in the NGS research laboratory, we
therefore share our experience of implementing a suc-
cessful LIMS.
Regarding further work, it is promising in our
schedule that we will provide a cloud-based LIMS so-
lution in our SolarGenomics genome sequencing big
data cloud platform, and open the service for more
genomics labs.
ACKNOWLEDGEMENTS
This work was partially supported by the Science
Foundation of Beijing Language and Culture Uni-
versity (supported by “the Fundamental Research
Funds for the Central Universities”) (15YJ030001,
14YBB12, 16YJ030001).
REFERENCES
Bath, T. G., Bozdag, S., Afzal, V., and Crowther, D.
(2011). Limsportal and Bonsailims: development of a
lab information management system for translational
medicine. In Source Code Biol Med.
Goodin, B., Poitras, C., and Begin, C. (2016). Mybatis. In
http://blog.mybatis.org/.
HEALTHINF 2017 - 10th International Conference on Health Informatics
332

Grimes, S. M. and Ji, H. P. (2014). Mendelims: a web-
based laboratory information management system for
clinical genome sequencing. In BMC Bioinformatics.
Hamosh, A., Scott, A. F., Amberger, J. S., Bocchini, C. A.,
and McKusick, V. A. (2015). Online mendelian in-
heritance in man (omim), a knowledgebase of human
genes and genetic disorders. In Nucleic Acids Re-
search.
Illumina (2016). Basespace Clarity LIMS. In
http://www.illumina.com/informatics/research/
sequencing-data-analysis-management/genologics-
lims.html.
Mindspec (2016). Autdb: a Genetic Database
for Autism Spectrum Disorders. In
http://www.mindspec.org/products/autdb/.
PerkinElmer (2016). Genesifter Lab Edition. In
http://www.geospiza.com/Products/LabEdition.shtml.
Pinero, J., Queralt-Rosinach, N., Bravo, A., Deu-Pons, J.,
Bauer-Mehren, A., Baron, M., Sanz, F., and Furlong,
L. I. (2015). Disgenet: a discovery platform for the
dynamical exploration of human diseases and their
genes. In Database (Oxford).
Pivotal (2016). Spring framework. In
http://projects.spring.io/spring-framework/.
Progeny (2016). Progeny LIMS. In
http://www.progenygenetics.com/lims/.
SolarGenomics (2016). Solargenomics genome sequencing
big data platform. In www.solargenomics.com.
Laboratory Information Management System for NGS Genomics Labs
333
